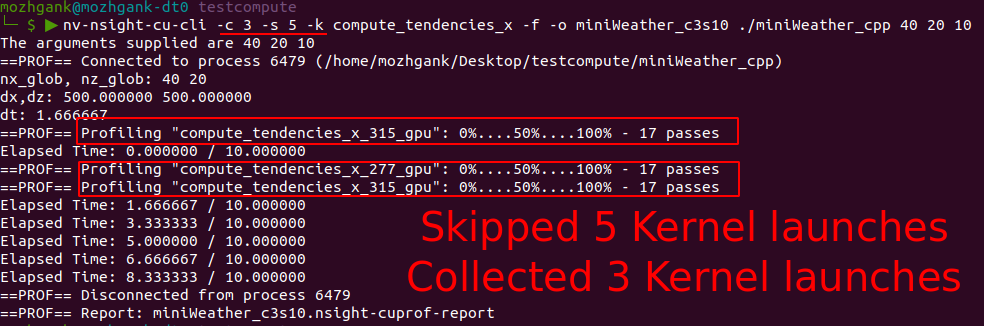
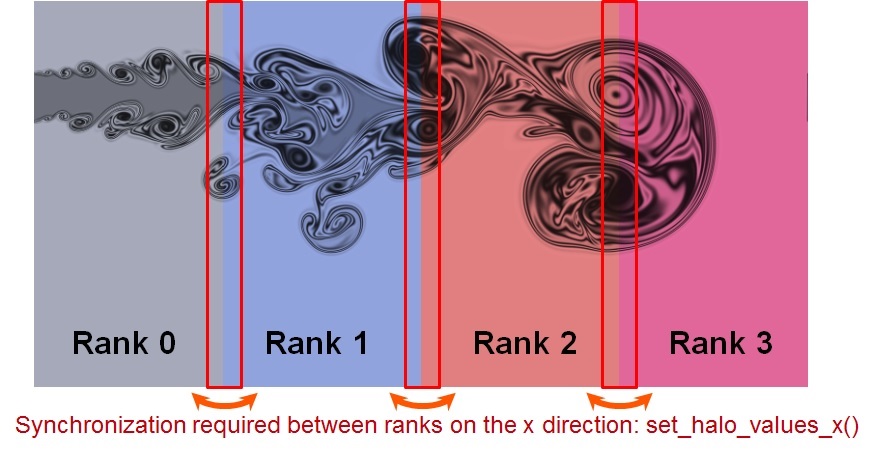
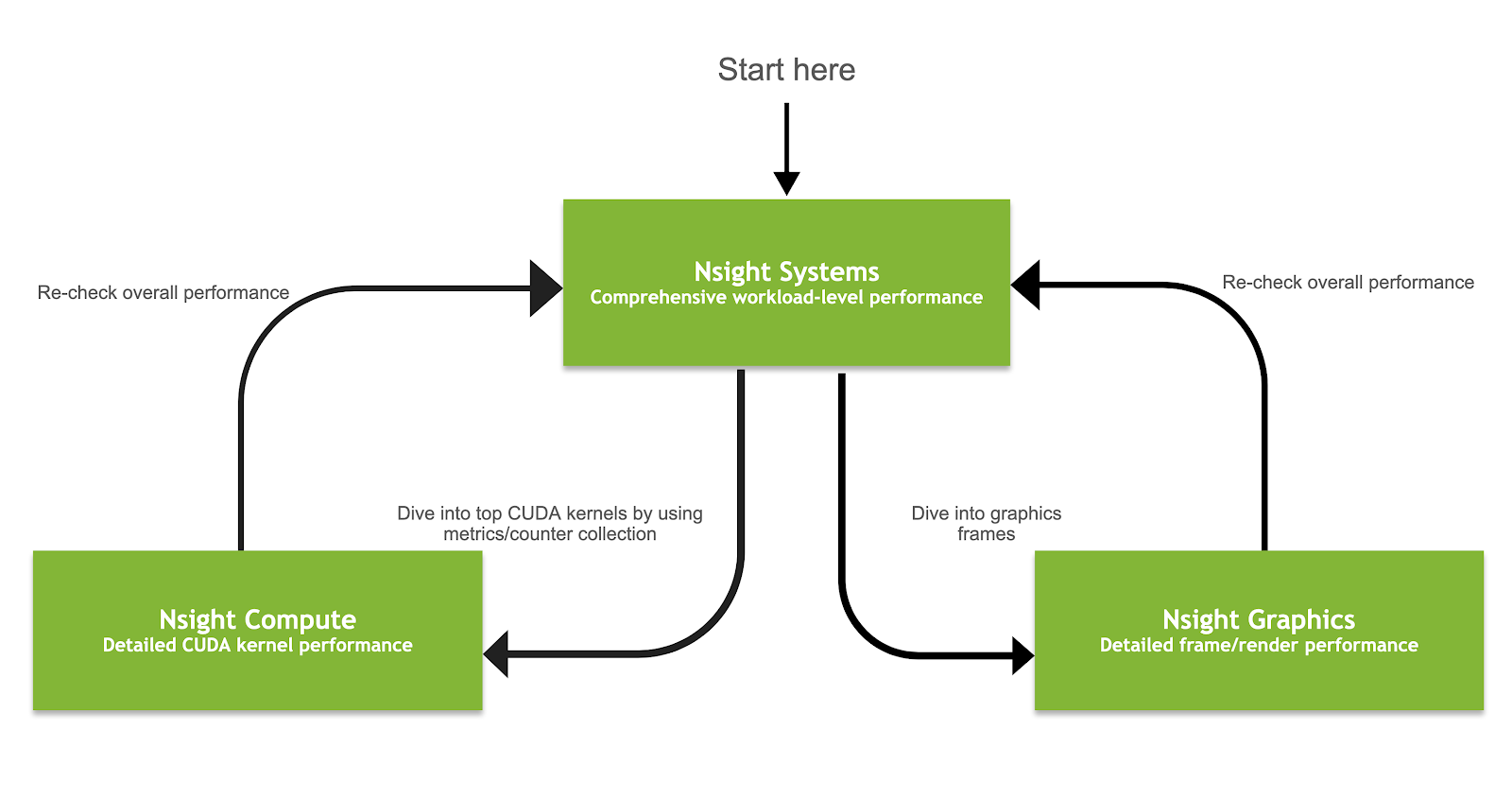
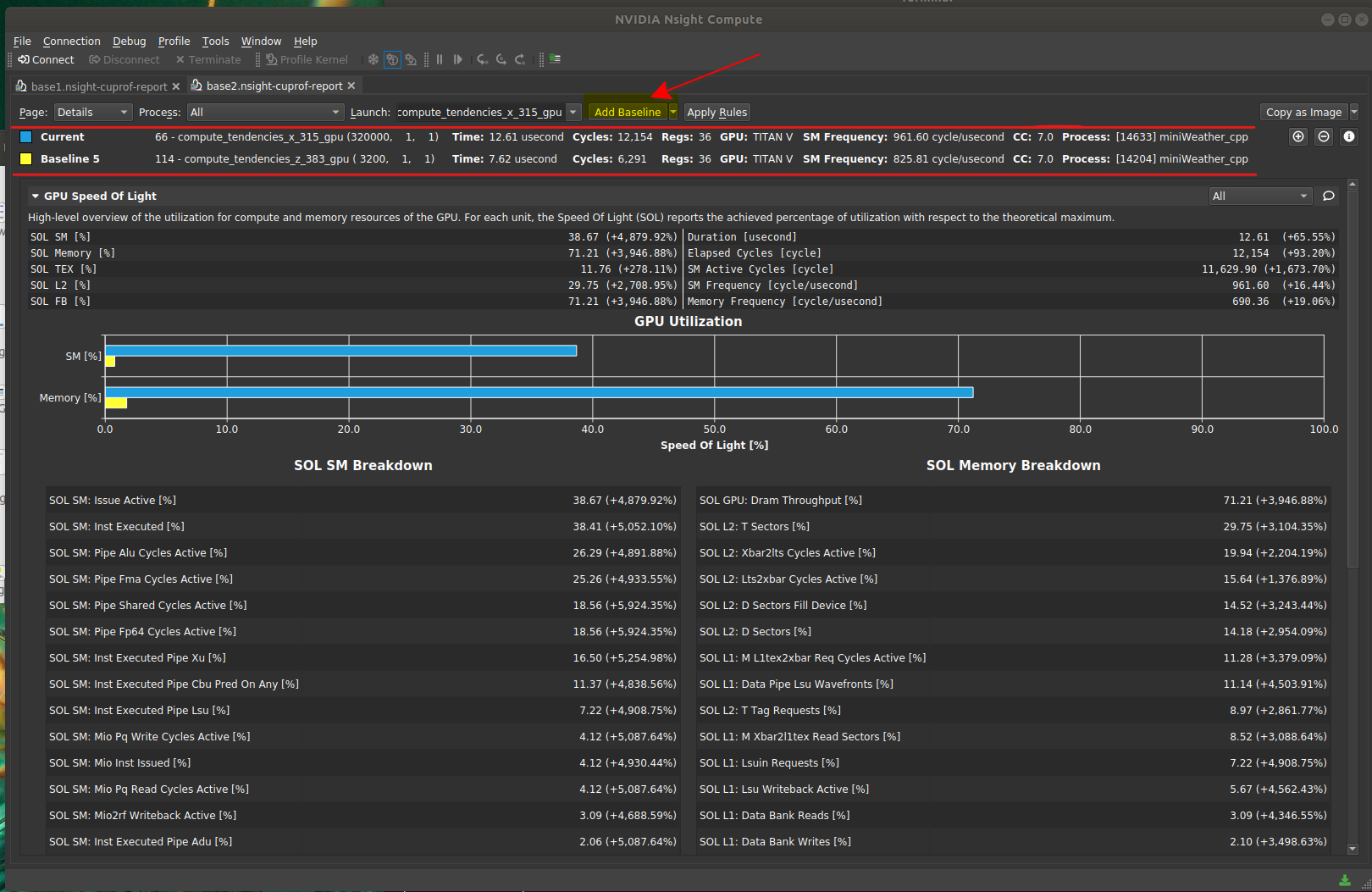
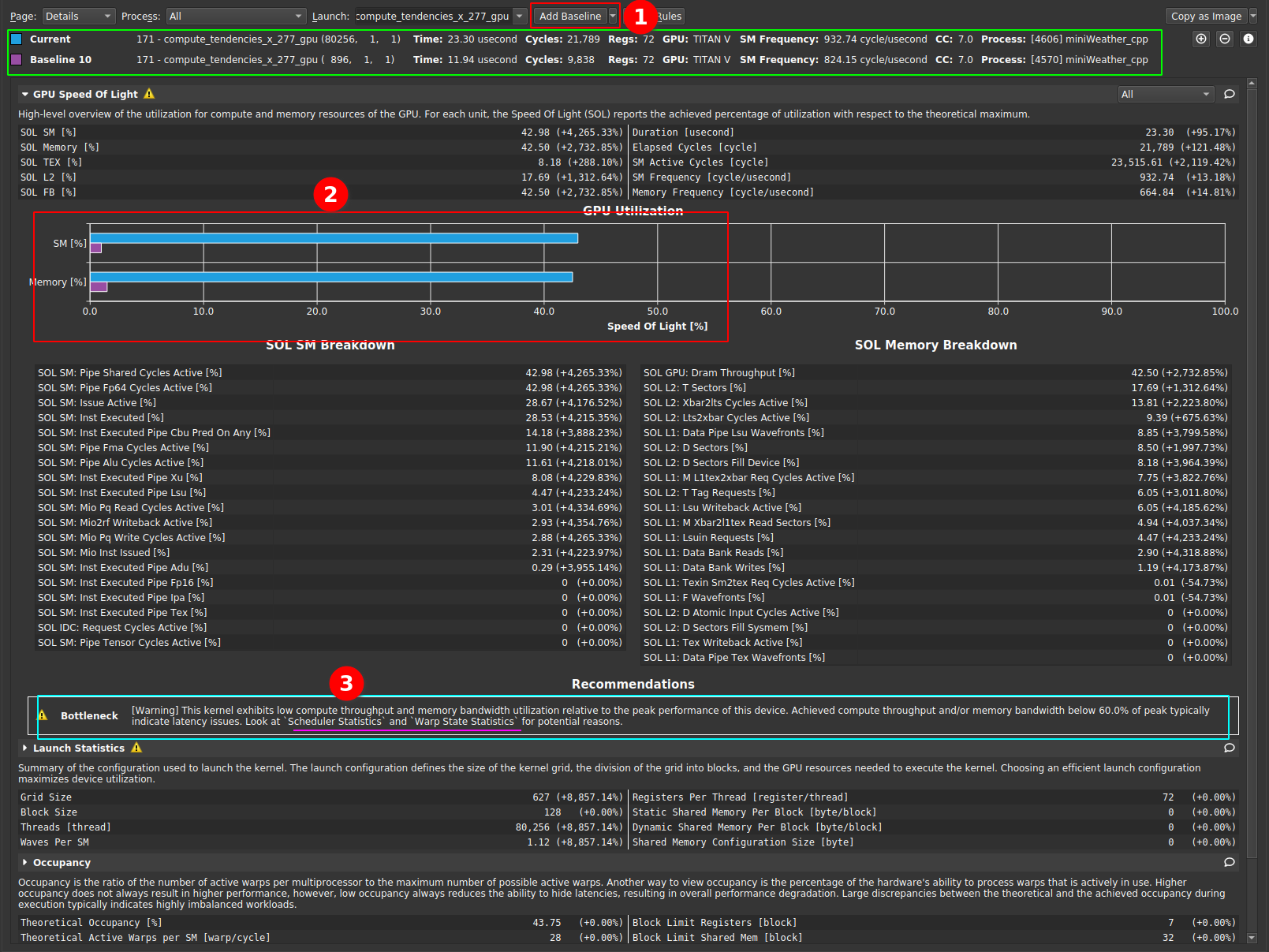
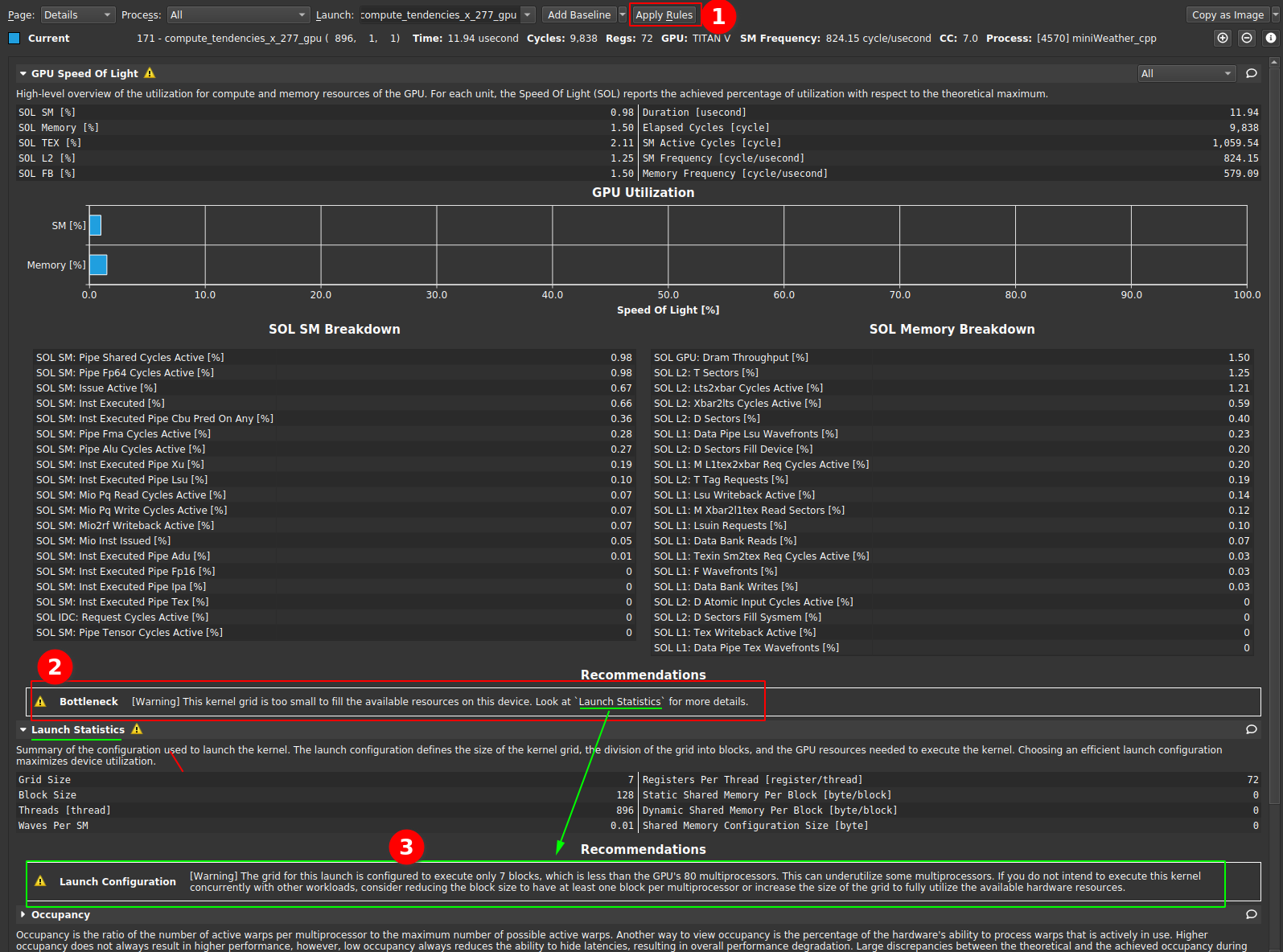
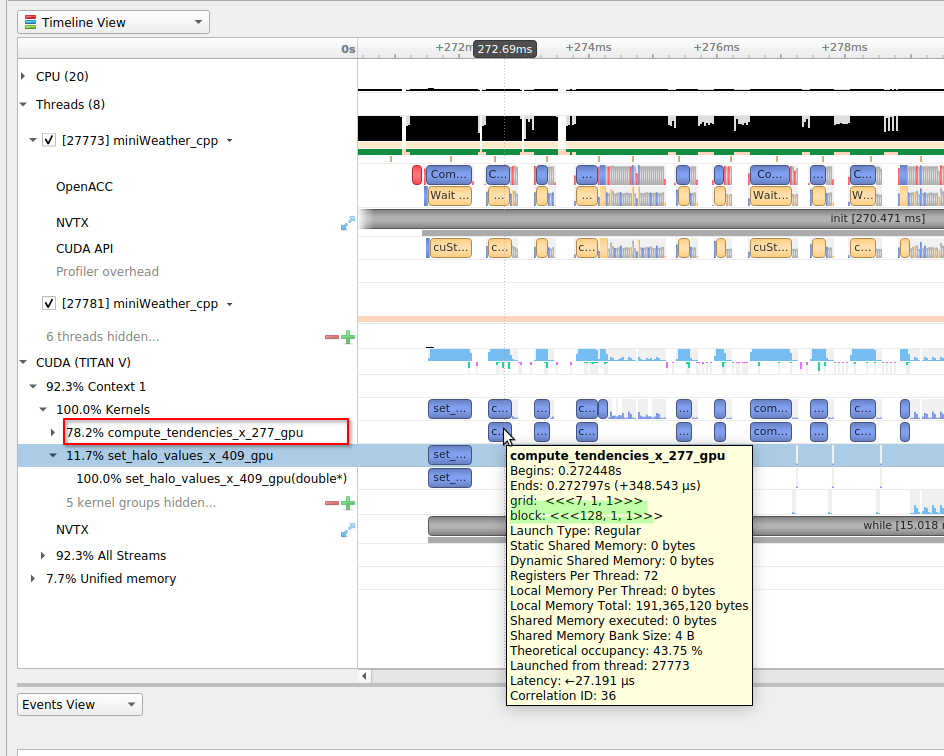
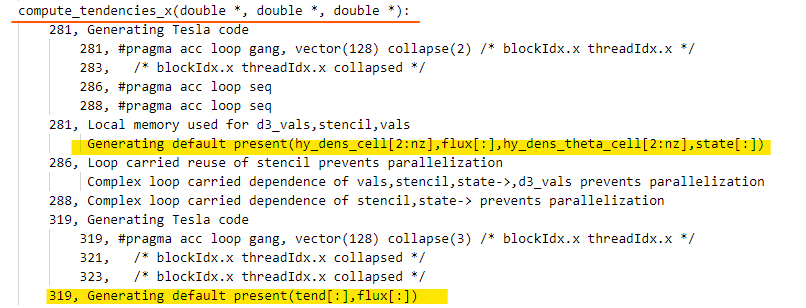
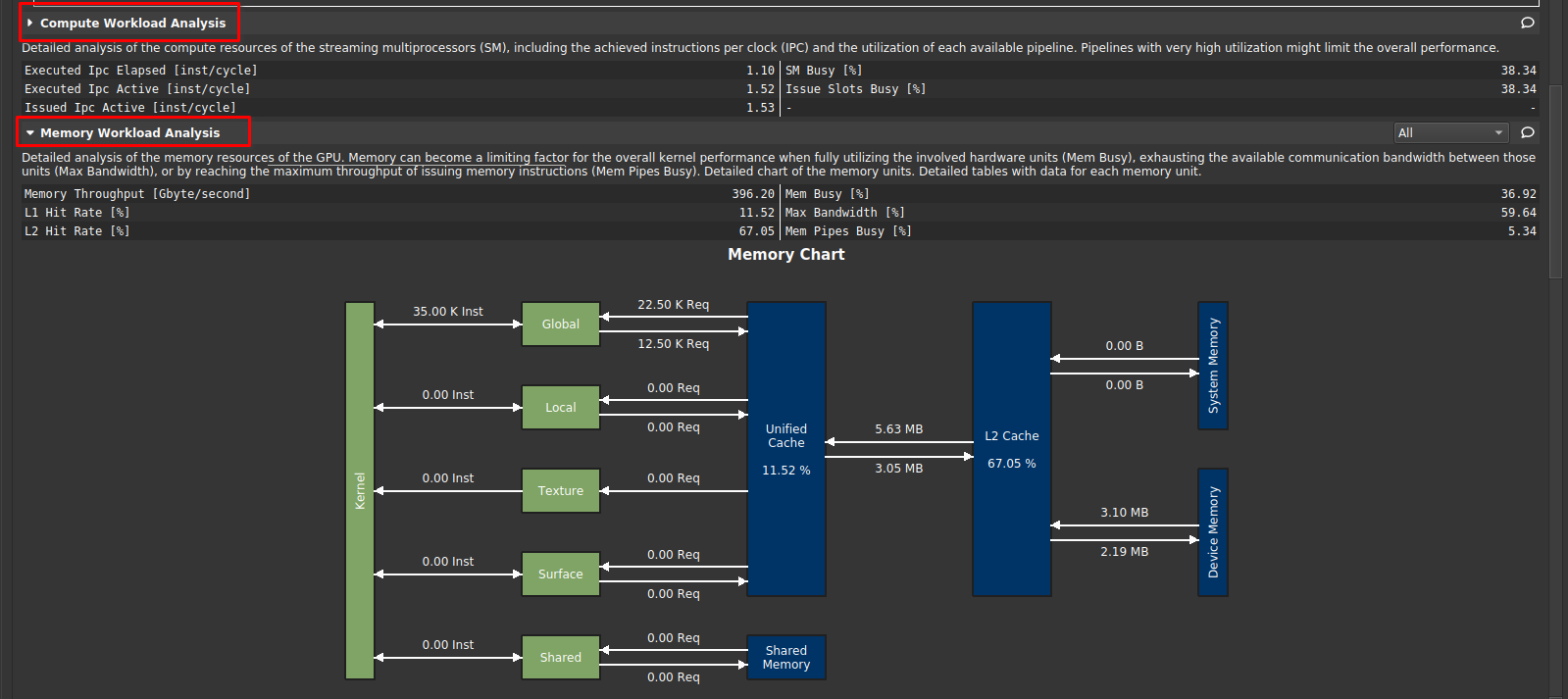
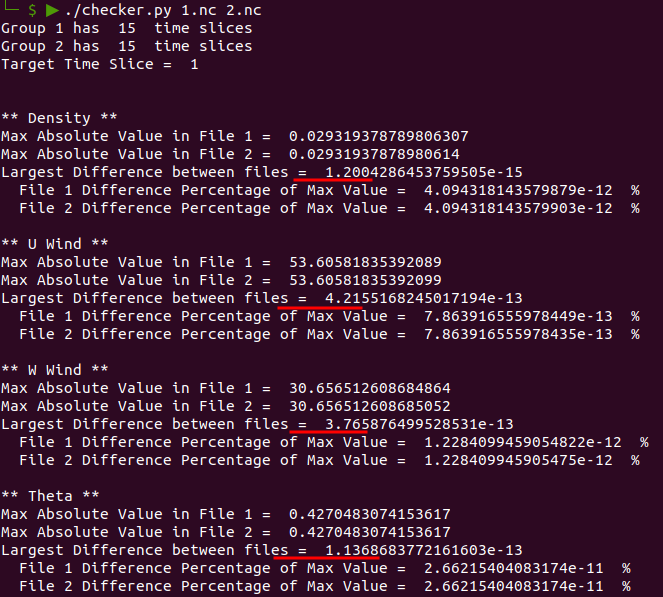
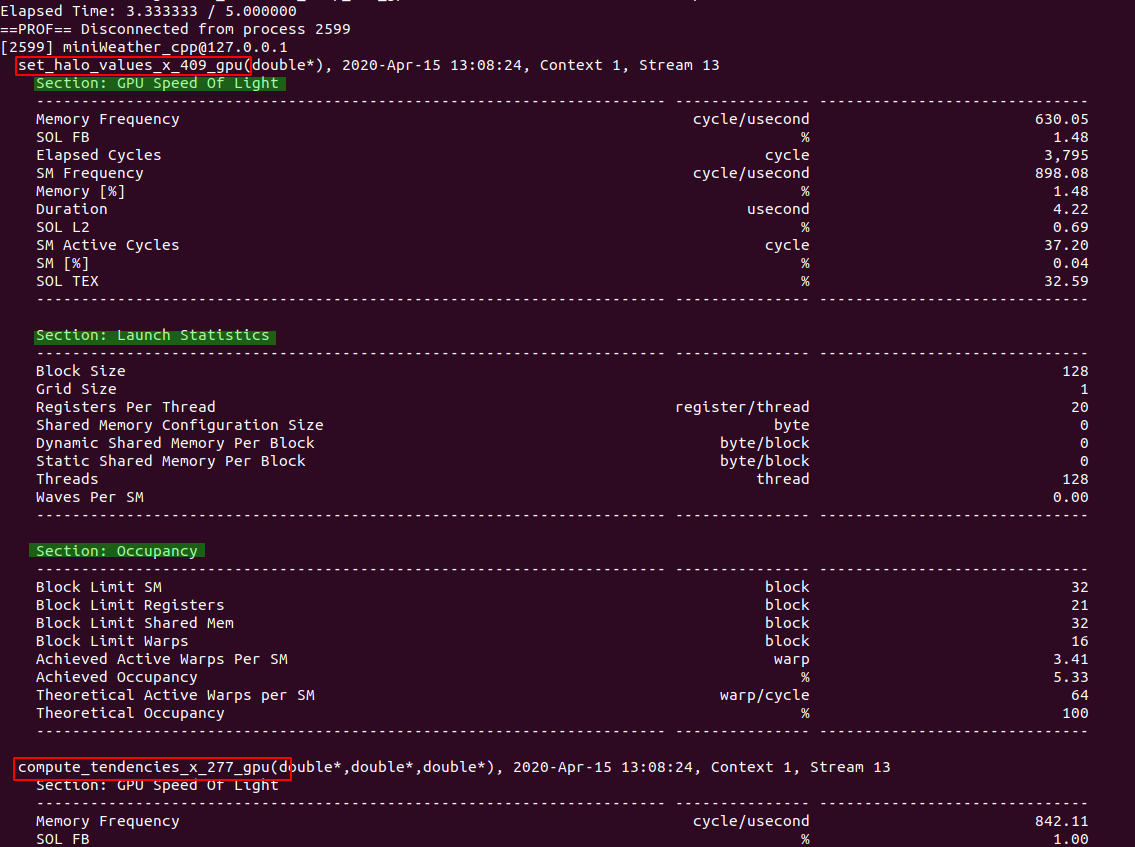
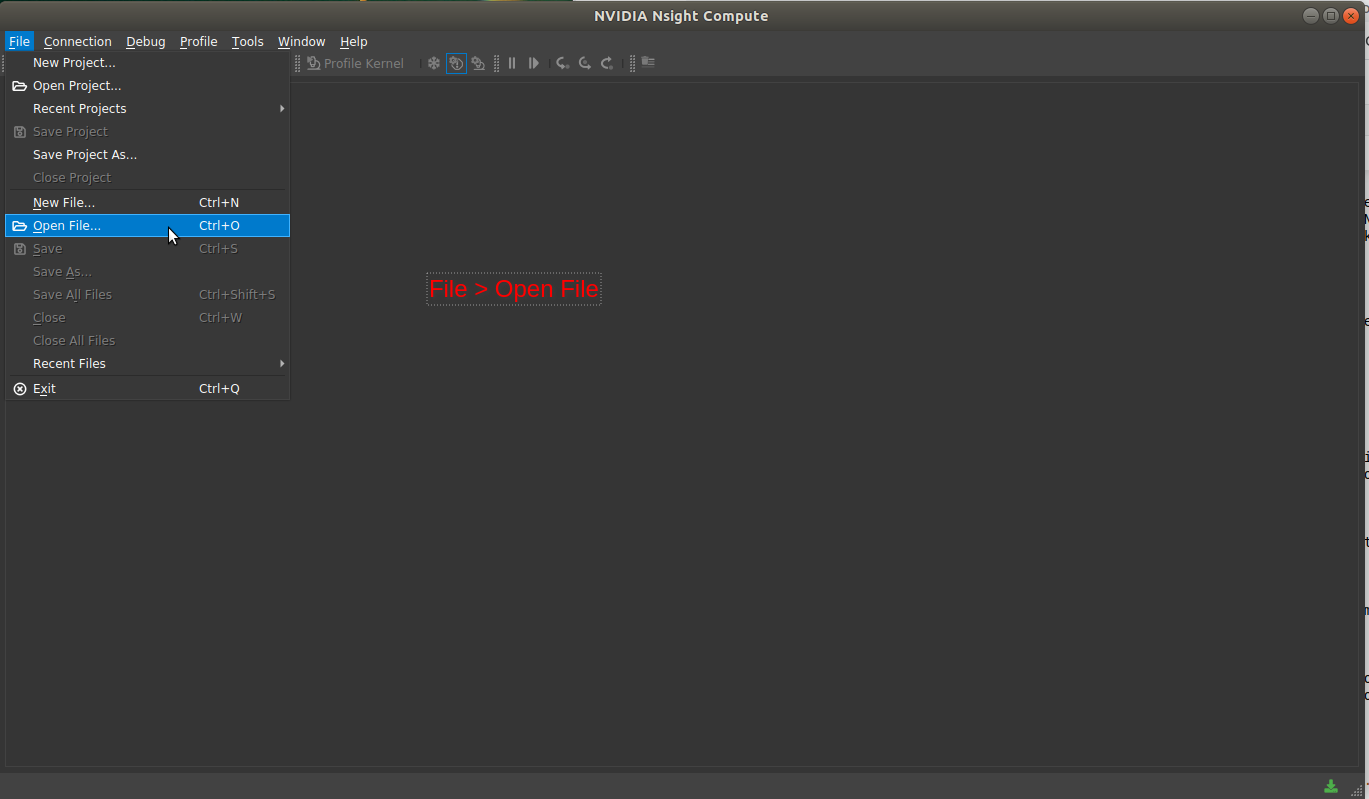
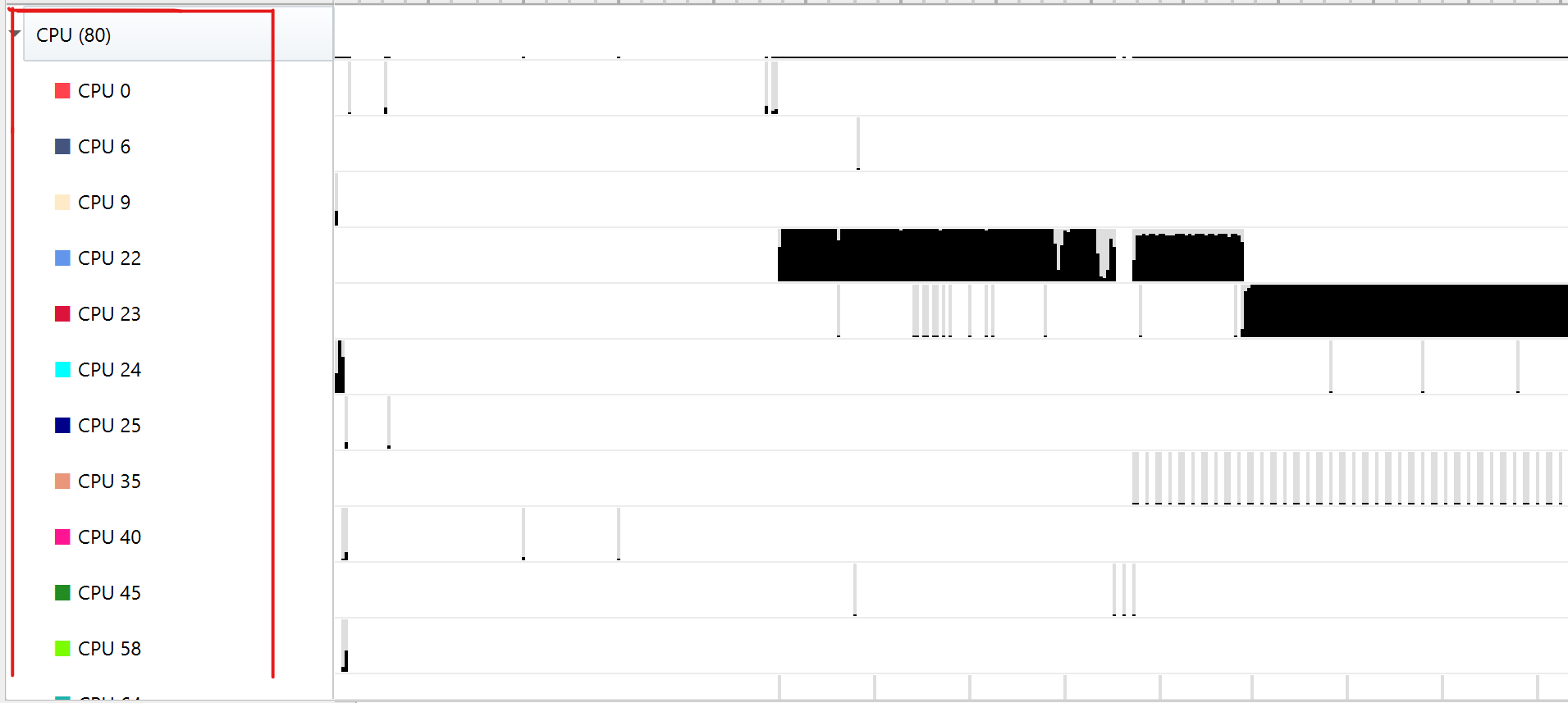
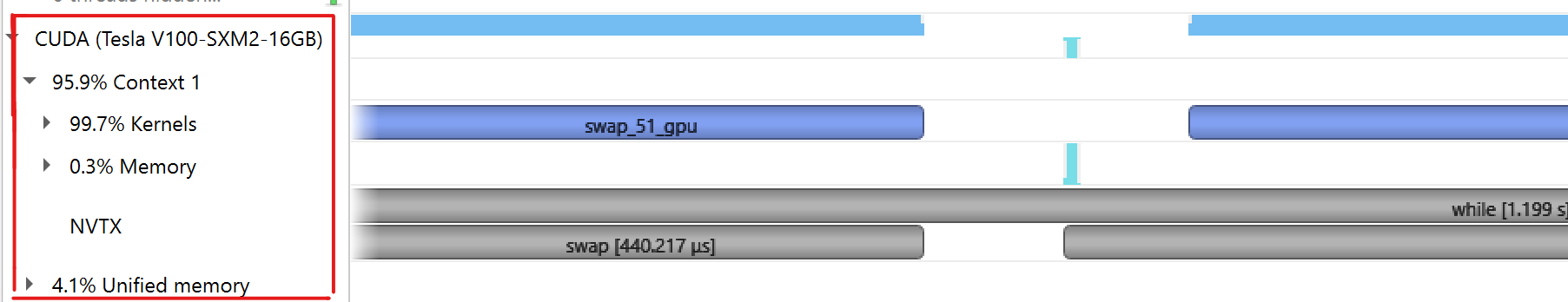
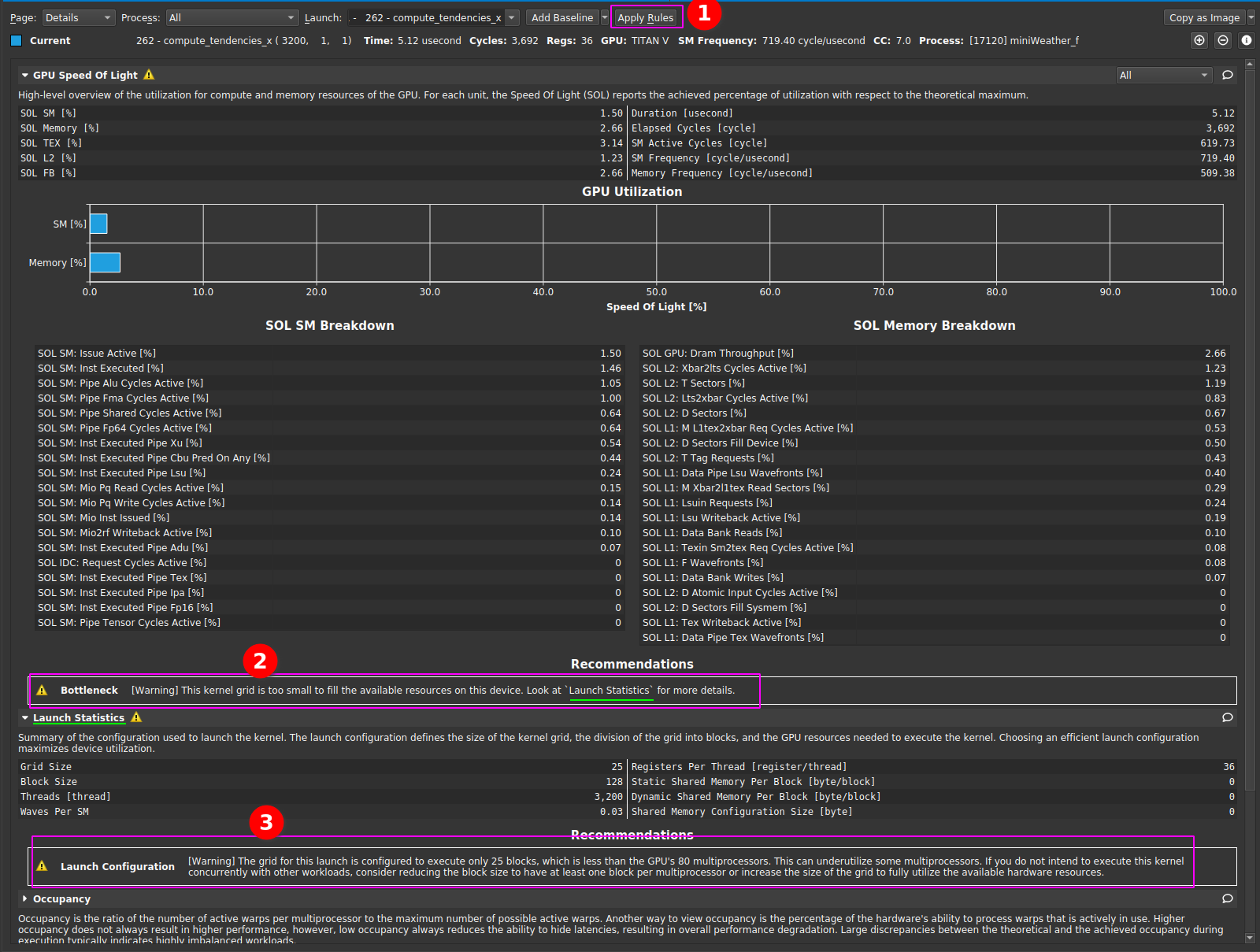
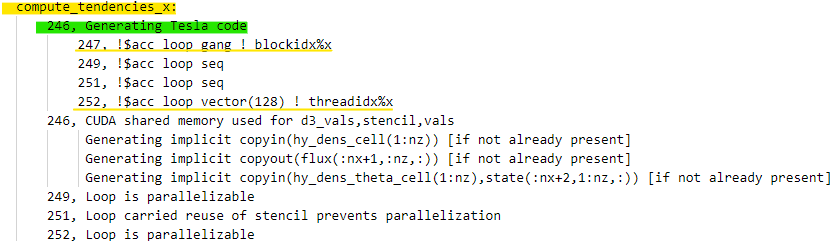
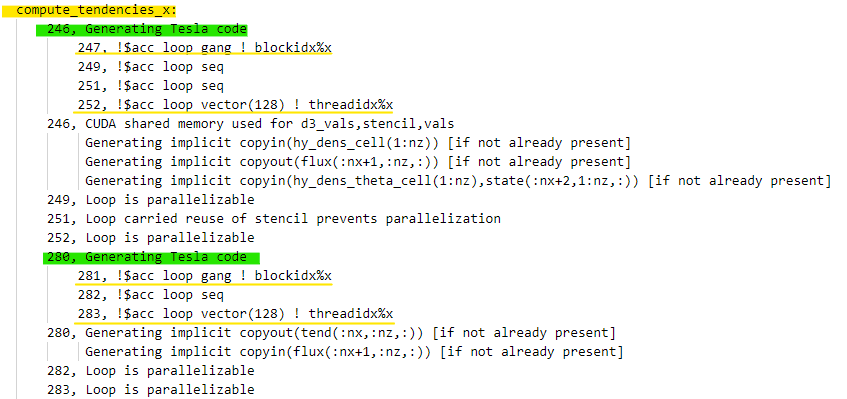
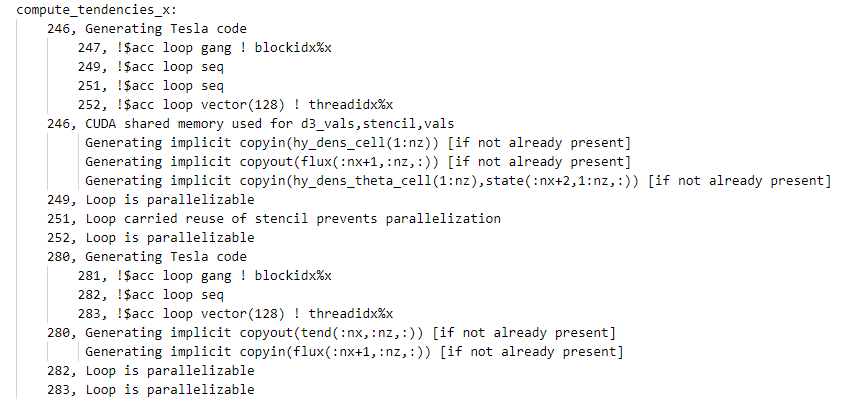
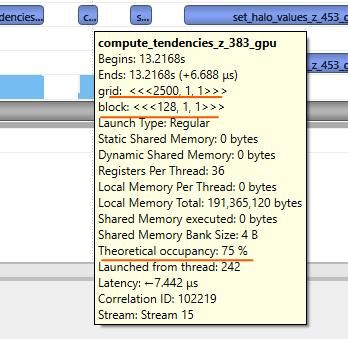
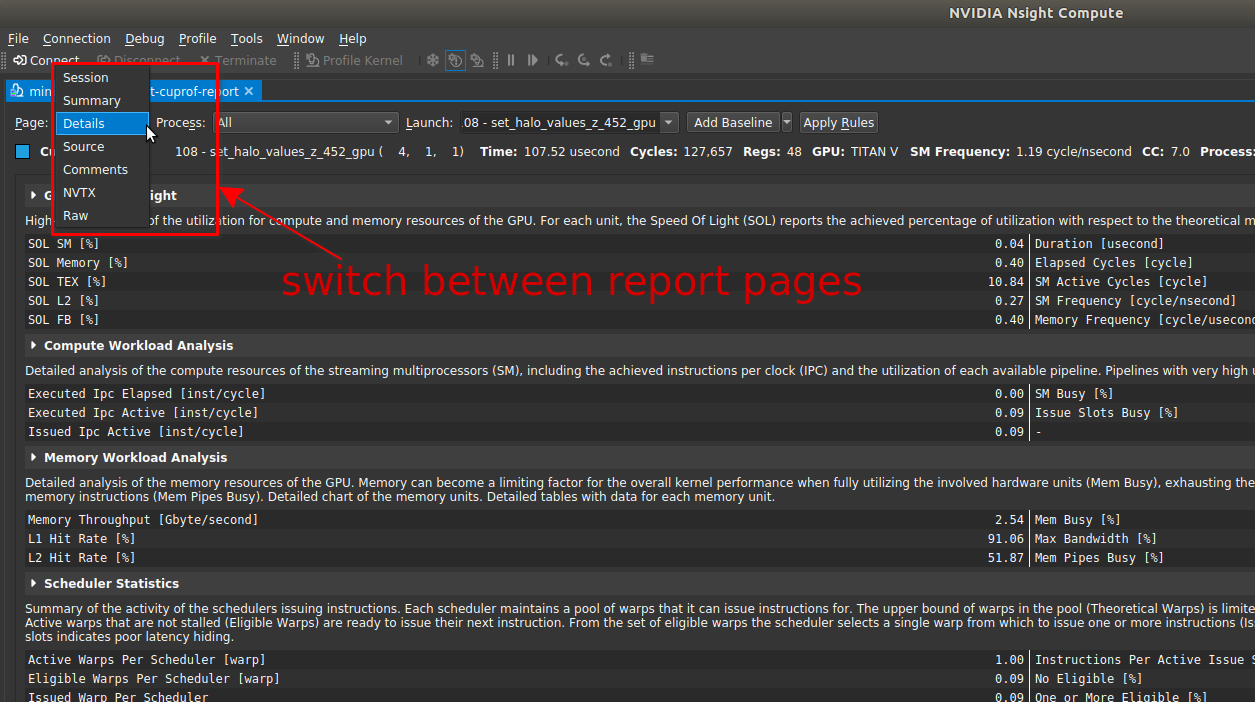
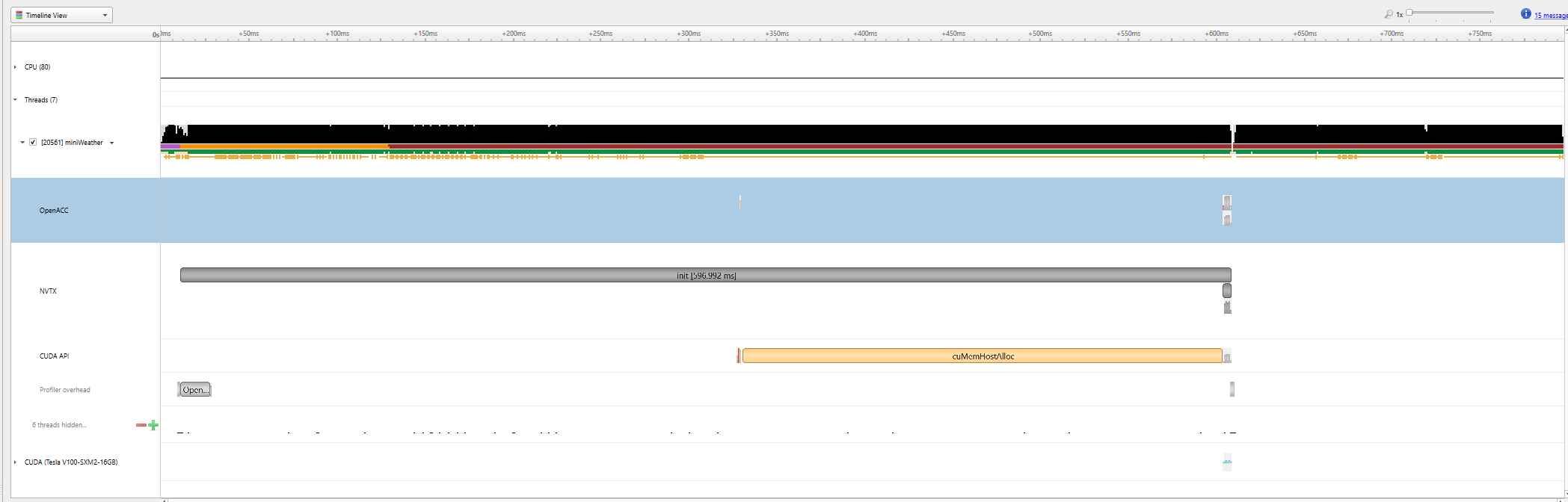
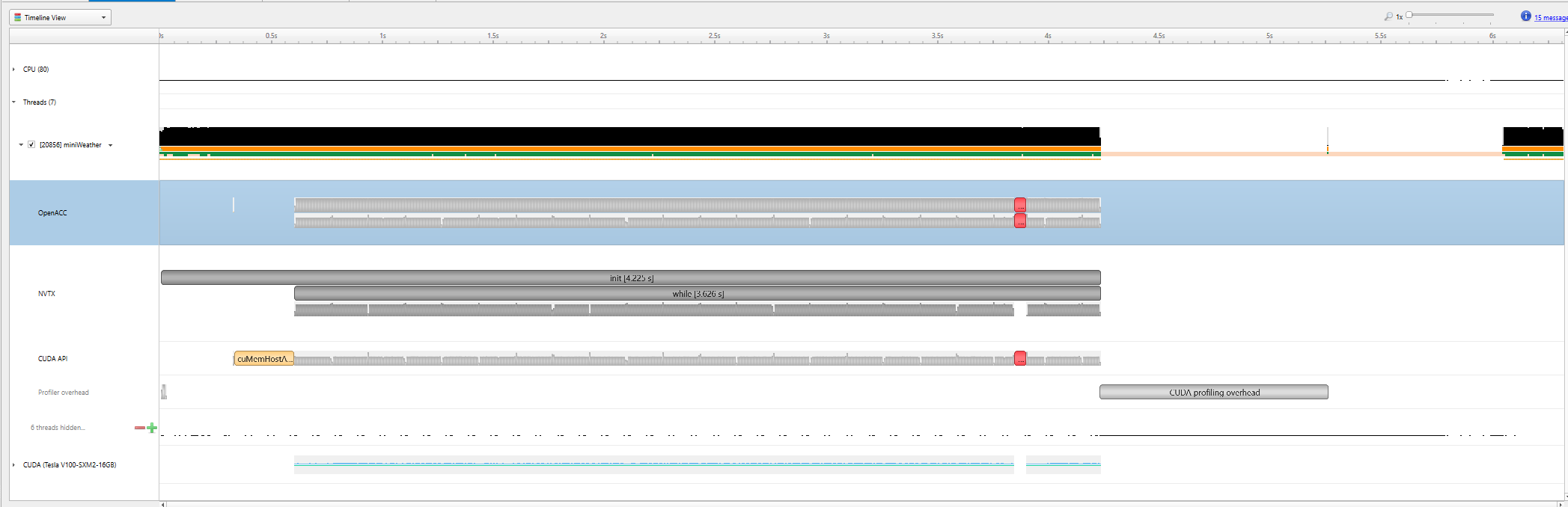
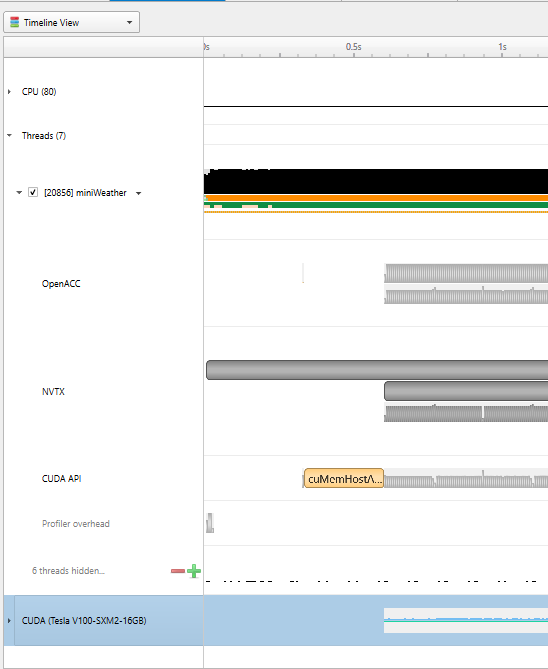
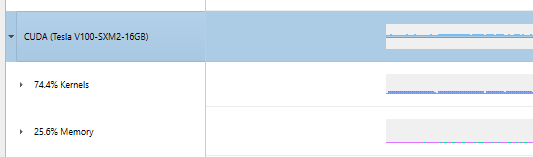
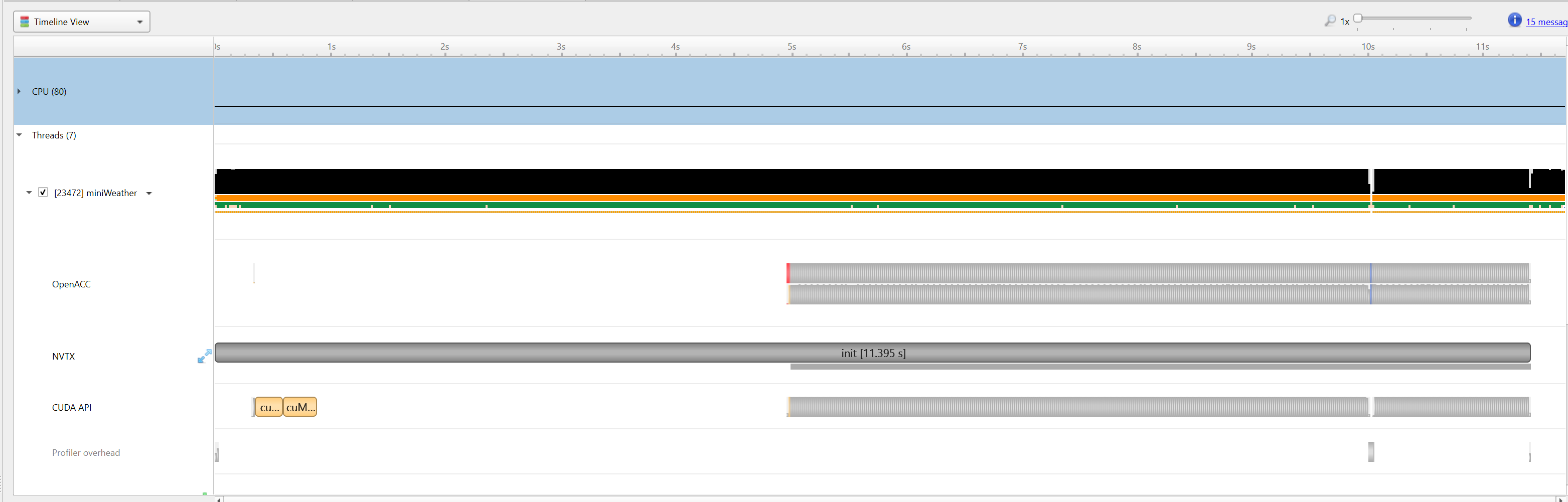
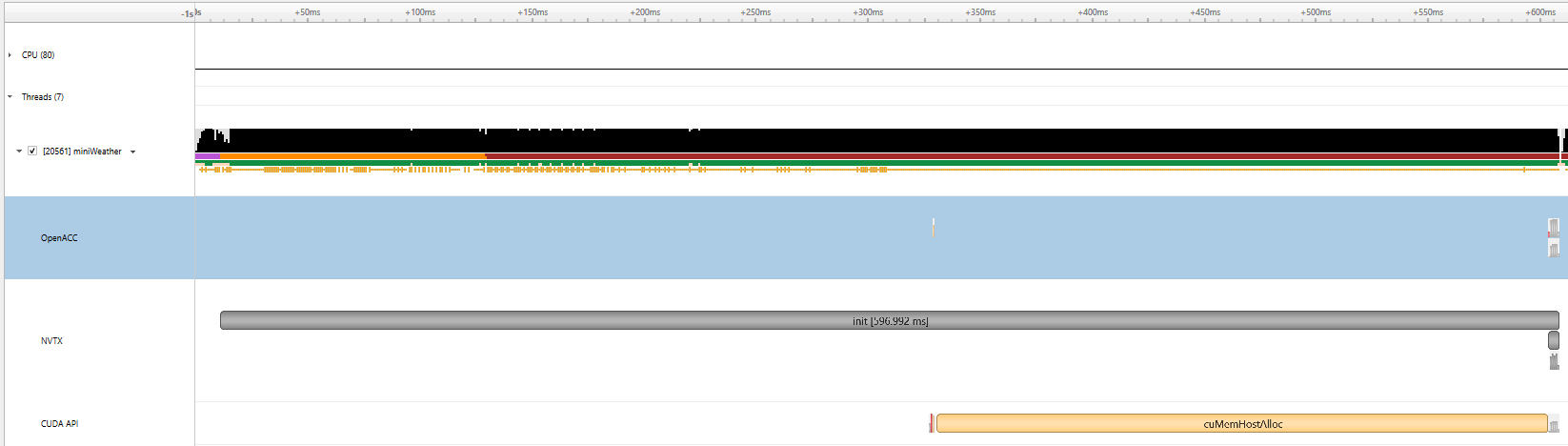
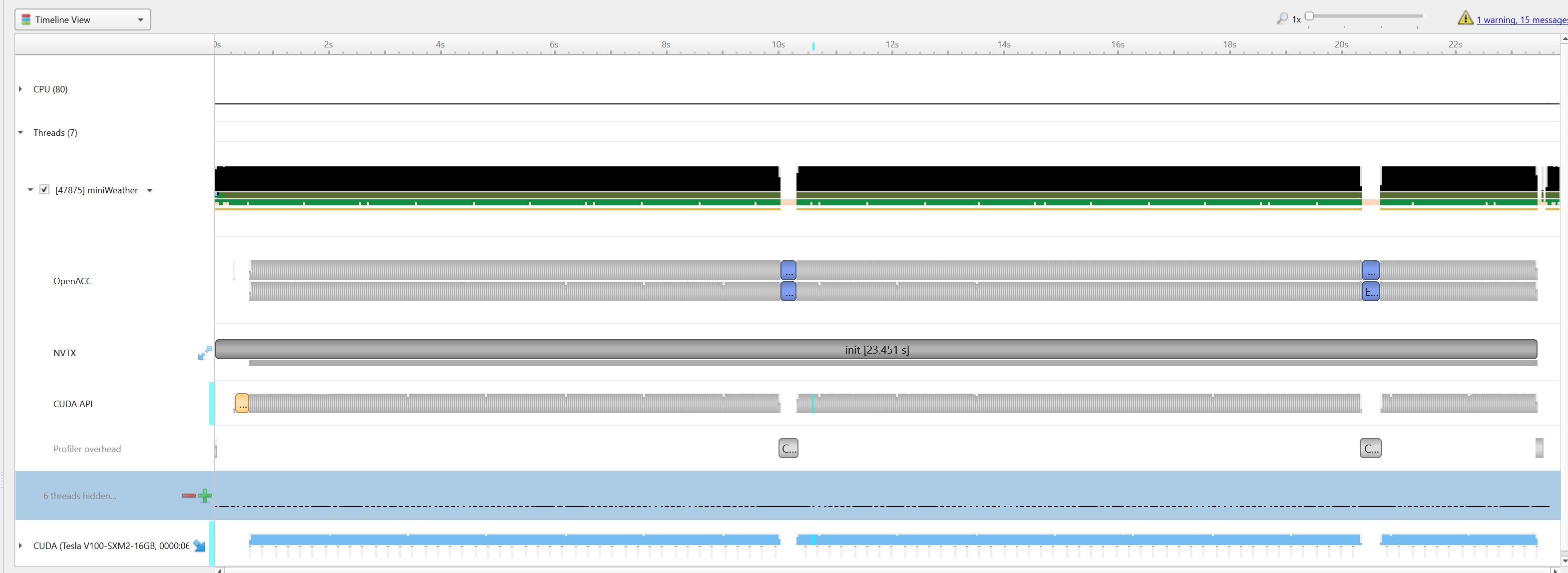
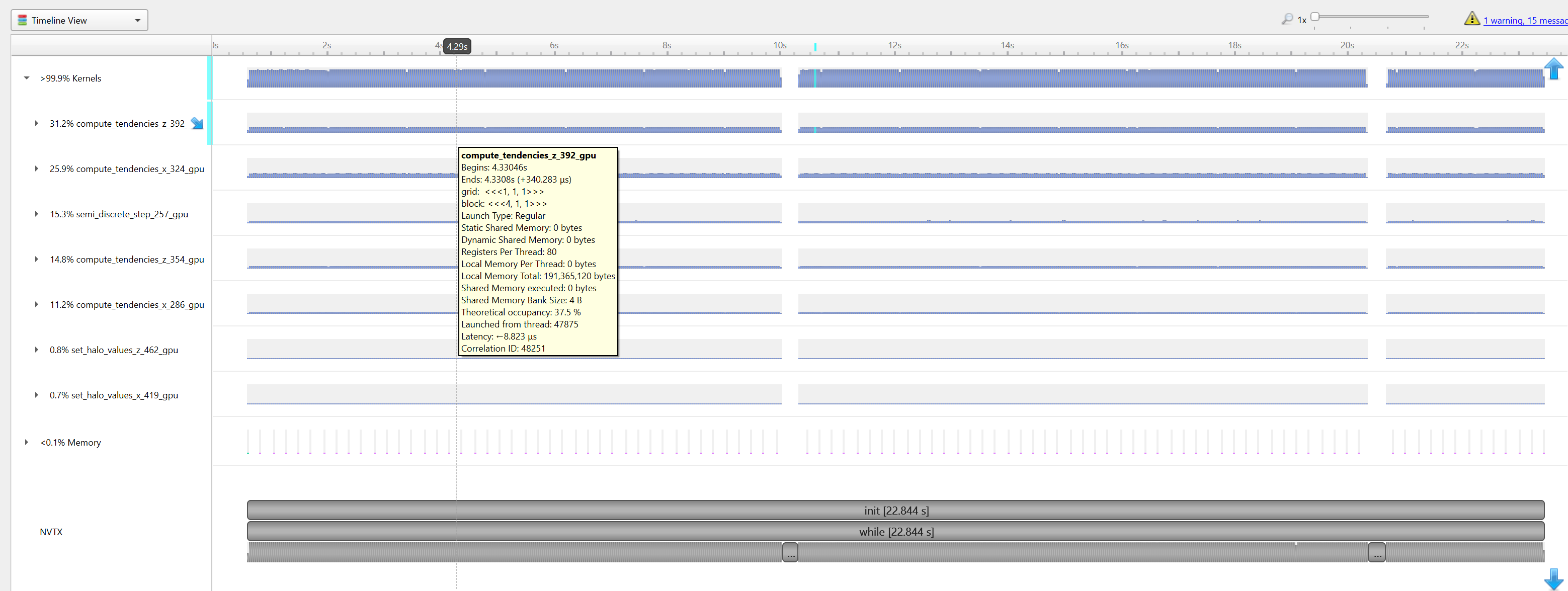
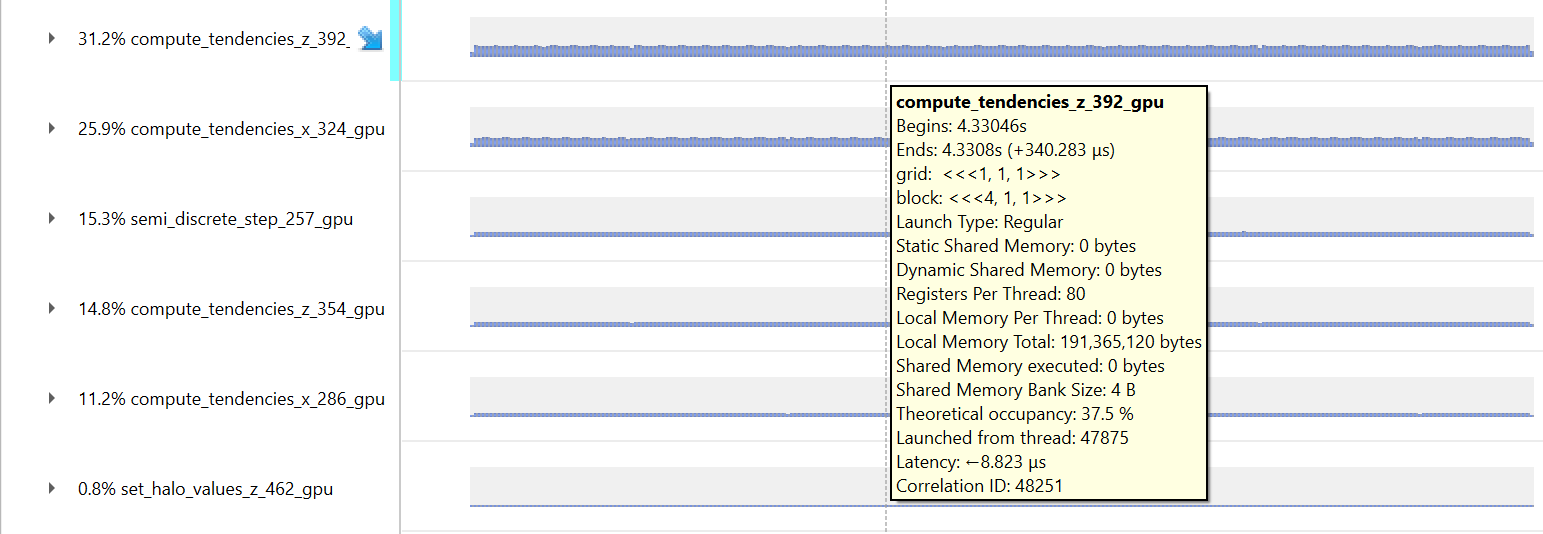
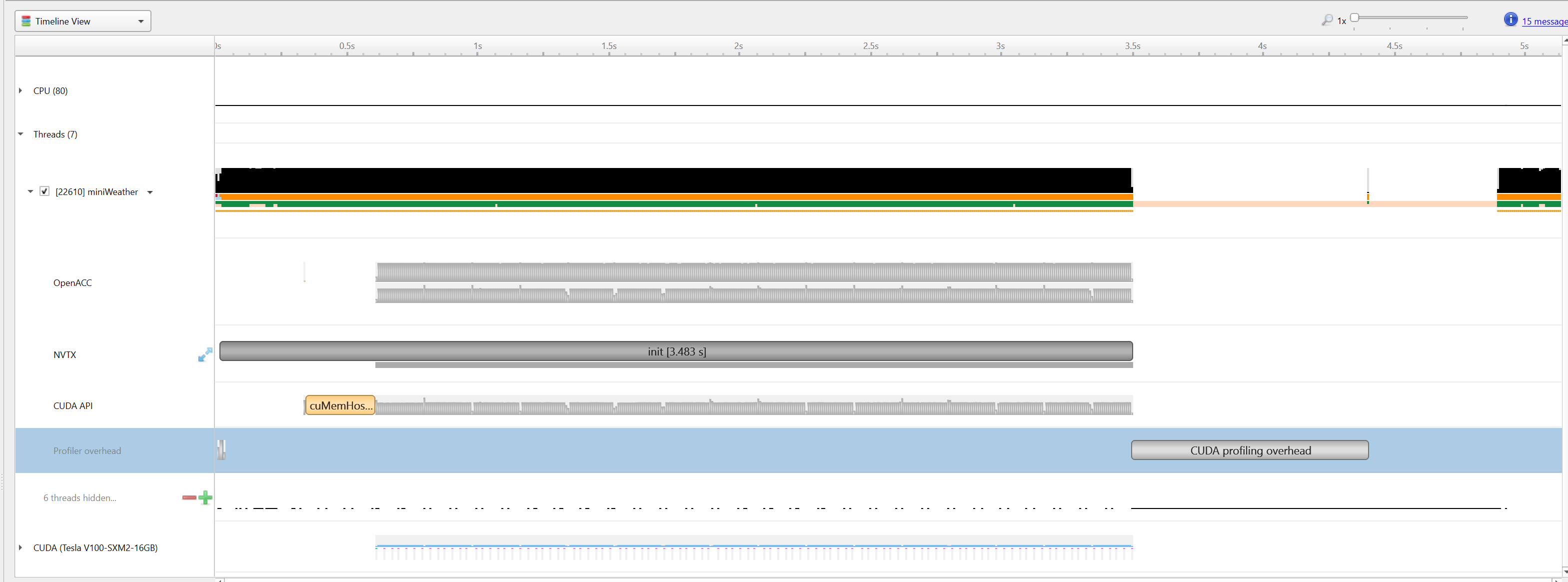
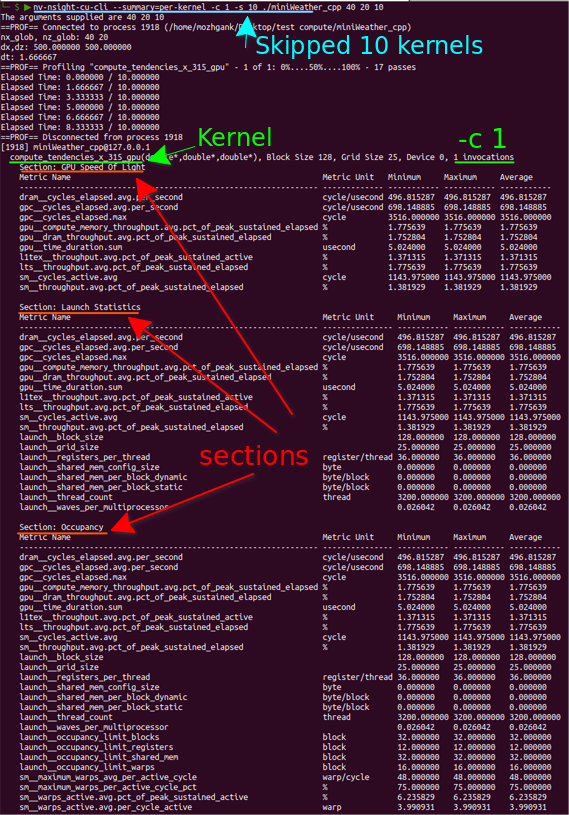
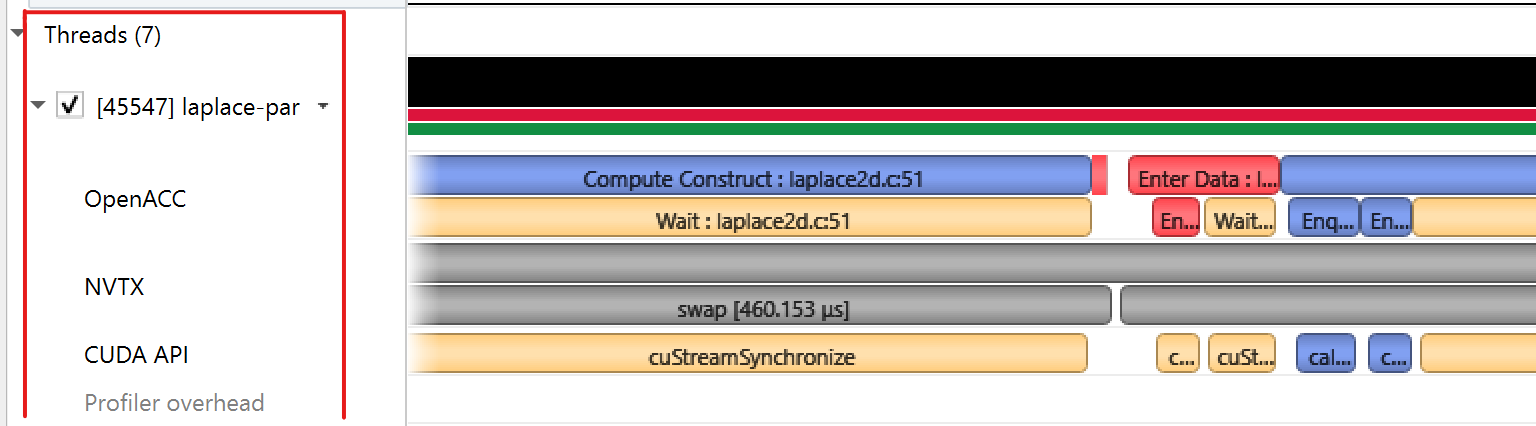
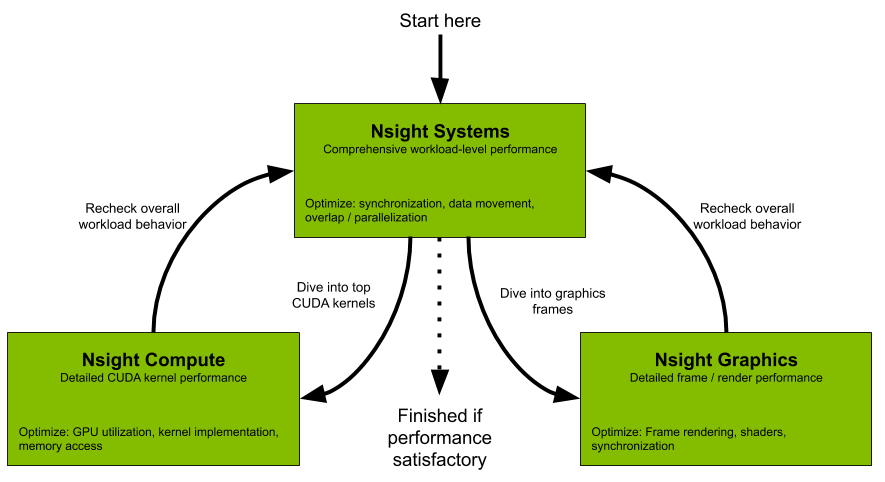
|
|
@@ -0,0 +1,766 @@
|
|
|
+//////////////////////////////////////////////////////////////////////////////////////////
|
|
|
+// miniWeather
|
|
|
+// Author: Matt Norman <normanmr@ornl.gov> , Oak Ridge National Laboratory
|
|
|
+// This code simulates dry, stratified, compressible, non-hydrostatic fluid flows
|
|
|
+// For documentation, please see the attached documentation in the "documentation" folder
|
|
|
+//////////////////////////////////////////////////////////////////////////////////////////
|
|
|
+
|
|
|
+#include <stdlib.h>
|
|
|
+#include <math.h>
|
|
|
+#include <stdio.h>
|
|
|
+#include <netcdf.h>
|
|
|
+#include <nvtx3/nvToolsExt.h>
|
|
|
+
|
|
|
+const double pi = 3.14159265358979323846264338327; //Pi
|
|
|
+const double grav = 9.8; //Gravitational acceleration (m / s^2)
|
|
|
+const double cp = 1004.; //Specific heat of dry air at constant pressure
|
|
|
+const double rd = 287.; //Dry air constant for equation of state (P=rho*rd*T)
|
|
|
+const double p0 = 1.e5; //Standard pressure at the surface in Pascals
|
|
|
+const double C0 = 27.5629410929725921310572974482; //Constant to translate potential temperature into pressure (P=C0*(rho*theta)**gamma)
|
|
|
+const double gamm = 1.40027894002789400278940027894; //gamma=cp/Rd , have to call this gamm because "gamma" is taken (I hate C so much)
|
|
|
+//Define domain and stability-related constants
|
|
|
+const double xlen = 2.e4; //Length of the domain in the x-direction (meters)
|
|
|
+const double zlen = 1.e4; //Length of the domain in the z-direction (meters)
|
|
|
+const double hv_beta = 0.25; //How strong to diffuse the solution: hv_beta \in [0:1]
|
|
|
+const double cfl = 1.50; //"Courant, Friedrichs, Lewy" number (for numerical stability)
|
|
|
+const double max_speed = 450; //Assumed maximum wave speed during the simulation (speed of sound + speed of wind) (meter / sec)
|
|
|
+const int hs = 2; //"Halo" size: number of cells needed for a full "stencil" of information for reconstruction
|
|
|
+const int sten_size = 4; //Size of the stencil used for interpolation
|
|
|
+
|
|
|
+//Parameters for indexing and flags
|
|
|
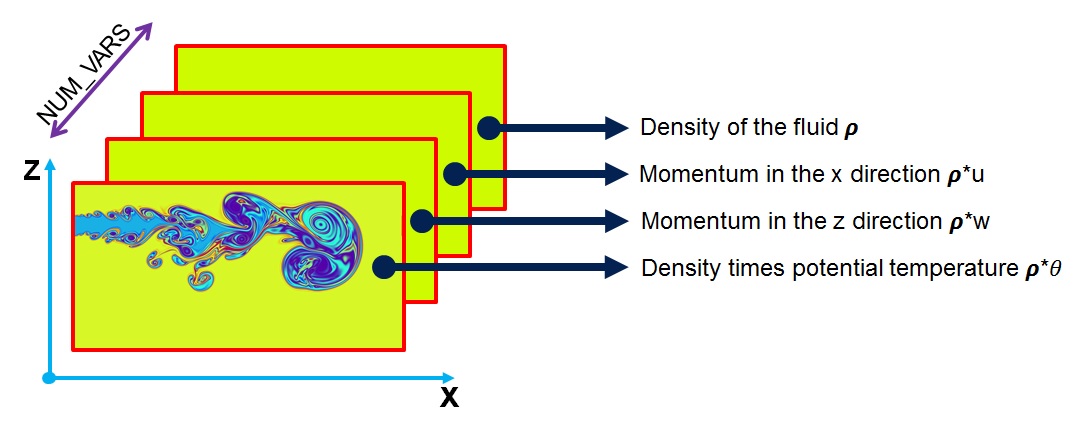
+const int NUM_VARS = 4; //Number of fluid state variables
|
|
|
+const int ID_DENS = 0; //index for density ("rho")
|
|
|
+const int ID_UMOM = 1; //index for momentum in the x-direction ("rho * u")
|
|
|
+const int ID_WMOM = 2; //index for momentum in the z-direction ("rho * w")
|
|
|
+const int ID_RHOT = 3; //index for density * potential temperature ("rho * theta")
|
|
|
+const int DIR_X = 1; //Integer constant to express that this operation is in the x-direction
|
|
|
+const int DIR_Z = 2; //Integer constant to express that this operation is in the z-direction
|
|
|
+
|
|
|
+const int nqpoints = 3;
|
|
|
+double qpoints[] = {0.112701665379258311482073460022E0, 0.500000000000000000000000000000E0, 0.887298334620741688517926539980E0};
|
|
|
+double qweights[] = {0.277777777777777777777777777779E0, 0.444444444444444444444444444444E0, 0.277777777777777777777777777779E0};
|
|
|
+
|
|
|
+///////////////////////////////////////////////////////////////////////////////////////
|
|
|
+// Variables that are initialized but remain static over the course of the simulation
|
|
|
+///////////////////////////////////////////////////////////////////////////////////////
|
|
|
+double sim_time; //total simulation time in seconds
|
|
|
+double output_freq; //frequency to perform output in seconds
|
|
|
+double dt; //Model time step (seconds)
|
|
|
+int nx, nz; //Number of local grid cells in the x- and z- dimensions
|
|
|
+double dx, dz; //Grid space length in x- and z-dimension (meters)
|
|
|
+int nx_glob, nz_glob; //Number of total grid cells in the x- and z- dimensions
|
|
|
+int i_beg, k_beg; //beginning index in the x- and z-directions
|
|
|
+int nranks, myrank; //my rank id
|
|
|
+int left_rank, right_rank; //Rank IDs that exist to my left and right in the global domain
|
|
|
+double *hy_dens_cell; //hydrostatic density (vert cell avgs). Dimensions: (1-hs:nz+hs)
|
|
|
+double *hy_dens_theta_cell; //hydrostatic rho*t (vert cell avgs). Dimensions: (1-hs:nz+hs)
|
|
|
+double *hy_dens_int; //hydrostatic density (vert cell interf). Dimensions: (1:nz+1)
|
|
|
+double *hy_dens_theta_int; //hydrostatic rho*t (vert cell interf). Dimensions: (1:nz+1)
|
|
|
+double *hy_pressure_int; //hydrostatic press (vert cell interf). Dimensions: (1:nz+1)
|
|
|
+
|
|
|
+///////////////////////////////////////////////////////////////////////////////////////
|
|
|
+// Variables that are dynamics over the course of the simulation
|
|
|
+///////////////////////////////////////////////////////////////////////////////////////
|
|
|
+double etime; //Elapsed model time
|
|
|
+double output_counter; //Helps determine when it's time to do output
|
|
|
+//Runtime variable arrays
|
|
|
+double *state; //Fluid state. Dimensions: (1-hs:nx+hs,1-hs:nz+hs,NUM_VARS)
|
|
|
+double *state_tmp; //Fluid state. Dimensions: (1-hs:nx+hs,1-hs:nz+hs,NUM_VARS)
|
|
|
+double *flux; //Cell interface fluxes. Dimensions: (nx+1,nz+1,NUM_VARS)
|
|
|
+double *tend; //Fluid state tendencies. Dimensions: (nx,nz,NUM_VARS)
|
|
|
+int num_out = 0; //The number of outputs performed so far
|
|
|
+int direction_switch = 1;
|
|
|
+
|
|
|
+//How is this not in the standard?!
|
|
|
+double dmin(double a, double b)
|
|
|
+{
|
|
|
+ if (a < b)
|
|
|
+ {
|
|
|
+ return a;
|
|
|
+ }
|
|
|
+ else
|
|
|
+ {
|
|
|
+ return b;
|
|
|
+ }
|
|
|
+};
|
|
|
+
|
|
|
+//Declaring the functions defined after "main"
|
|
|
+void init();
|
|
|
+void finalize();
|
|
|
+void injection(double x, double z, double &r, double &u, double &w, double &t, double &hr, double &ht);
|
|
|
+void hydro_const_theta(double z, double &r, double &t);
|
|
|
+void output(double *state, double etime);
|
|
|
+void ncwrap(int ierr, int line);
|
|
|
+void perform_timestep(double *state, double *state_tmp, double *flux, double *tend, double dt);
|
|
|
+void semi_discrete_step(double *state_init, double *state_forcing, double *state_out, double dt, int dir, double *flux, double *tend);
|
|
|
+void compute_tendencies_x(double *state, double *flux, double *tend);
|
|
|
+void compute_tendencies_z(double *state, double *flux, double *tend);
|
|
|
+void set_halo_values_x(double *state);
|
|
|
+void set_halo_values_z(double *state);
|
|
|
+
|
|
|
+///////////////////////////////////////////////////////////////////////////////////////
|
|
|
+// THE MAIN PROGRAM STARTS HERE
|
|
|
+///////////////////////////////////////////////////////////////////////////////////////
|
|
|
+int main(int argc, char **argv)
|
|
|
+{
|
|
|
+ ///////////////////////////////////////////////////////////////////////////////////////
|
|
|
+ // BEGIN USER-CONFIGURABLE PARAMETERS
|
|
|
+ ///////////////////////////////////////////////////////////////////////////////////////
|
|
|
+ //The x-direction length is twice as long as the z-direction length
|
|
|
+ //So, you'll want to have nx_glob be twice as large as nz_glob
|
|
|
+ nx_glob = 400; //Number of total cells in the x-dirction
|
|
|
+ nz_glob = 200; //Number of total cells in the z-dirction
|
|
|
+ sim_time = 1500; //How many seconds to run the simulation
|
|
|
+ output_freq = 100; //How frequently to output data to file (in seconds)
|
|
|
+ ///////////////////////////////////////////////////////////////////////////////////////
|
|
|
+ // END USER-CONFIGURABLE PARAMETERS
|
|
|
+ ///////////////////////////////////////////////////////////////////////////////////////
|
|
|
+
|
|
|
+ if (argc == 4)
|
|
|
+ {
|
|
|
+ printf("The arguments supplied are %s %s %s\n", argv[1], argv[2], argv[3]);
|
|
|
+ nx_glob = atoi(argv[1]);
|
|
|
+ nz_glob = atoi(argv[2]);
|
|
|
+ sim_time = atoi(argv[3]);
|
|
|
+ }
|
|
|
+ else
|
|
|
+ {
|
|
|
+ printf("Using default values ...\n");
|
|
|
+ }
|
|
|
+
|
|
|
+ nvtxRangePushA("Total");
|
|
|
+ init();
|
|
|
+
|
|
|
+ //Output the initial state
|
|
|
+ output(state, etime);
|
|
|
+
|
|
|
+ ////////////////////////////////////////////////////
|
|
|
+ // MAIN TIME STEP LOOP
|
|
|
+ ////////////////////////////////////////////////////
|
|
|
+
|
|
|
+ nvtxRangePushA("while");
|
|
|
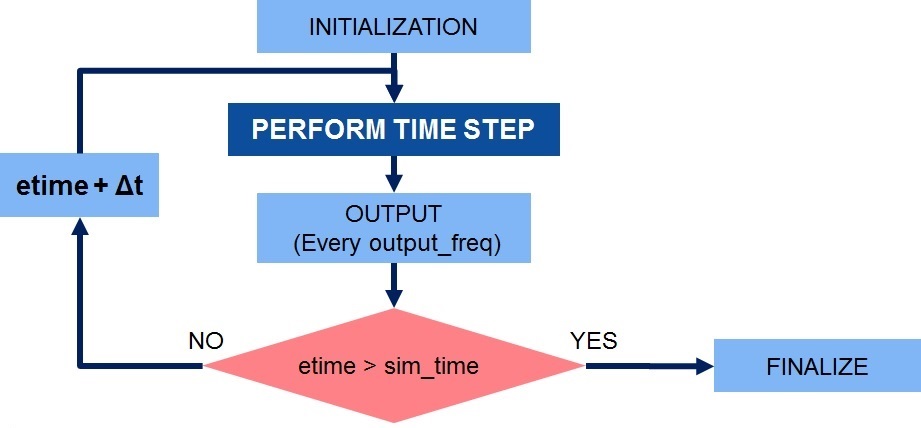
+ while (etime < sim_time)
|
|
|
+ {
|
|
|
+ //If the time step leads to exceeding the simulation time, shorten it for the last step
|
|
|
+ if (etime + dt > sim_time)
|
|
|
+ {
|
|
|
+ dt = sim_time - etime;
|
|
|
+ }
|
|
|
+
|
|
|
+ //Perform a single time step
|
|
|
+ nvtxRangePushA("perform_timestep");
|
|
|
+ perform_timestep(state, state_tmp, flux, tend, dt);
|
|
|
+ nvtxRangePop();
|
|
|
+
|
|
|
+ //Inform the user
|
|
|
+
|
|
|
+ printf("Elapsed Time: %lf / %lf\n", etime, sim_time);
|
|
|
+
|
|
|
+ //Update the elapsed time and output counter
|
|
|
+ etime = etime + dt;
|
|
|
+ output_counter = output_counter + dt;
|
|
|
+ //If it's time for output, reset the counter, and do output
|
|
|
+
|
|
|
+ if (output_counter >= output_freq)
|
|
|
+ {
|
|
|
+ output_counter = output_counter - output_freq;
|
|
|
+
|
|
|
+ output(state, etime);
|
|
|
+ }
|
|
|
+ }
|
|
|
+ nvtxRangePop();
|
|
|
+
|
|
|
+ finalize();
|
|
|
+ nvtxRangePop();
|
|
|
+}
|
|
|
+
|
|
|
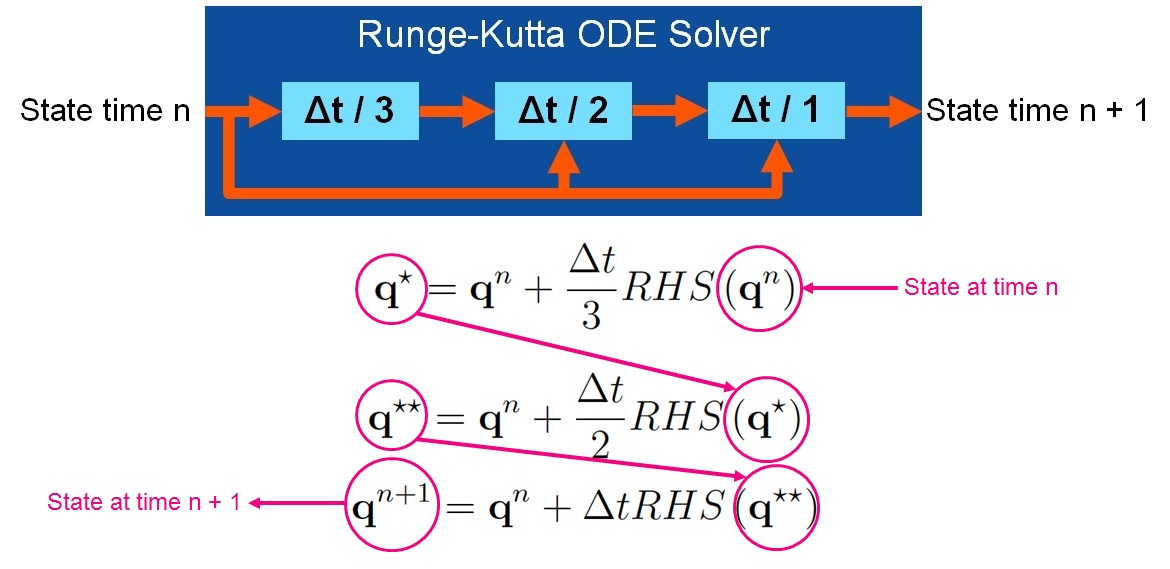
+//Performs a single dimensionally split time step using a simple low-storate three-stage Runge-Kutta time integrator
|
|
|
+//The dimensional splitting is a second-order-accurate alternating Strang splitting in which the
|
|
|
+//order of directions is alternated each time step.
|
|
|
+//The Runge-Kutta method used here is defined as follows:
|
|
|
+// q* = q[n] + dt/3 * rhs(q[n])
|
|
|
+// q** = q[n] + dt/2 * rhs(q* )
|
|
|
+// q[n+1] = q[n] + dt/1 * rhs(q** )
|
|
|
+void perform_timestep(double *state, double *state_tmp, double *flux, double *tend, double dt)
|
|
|
+{
|
|
|
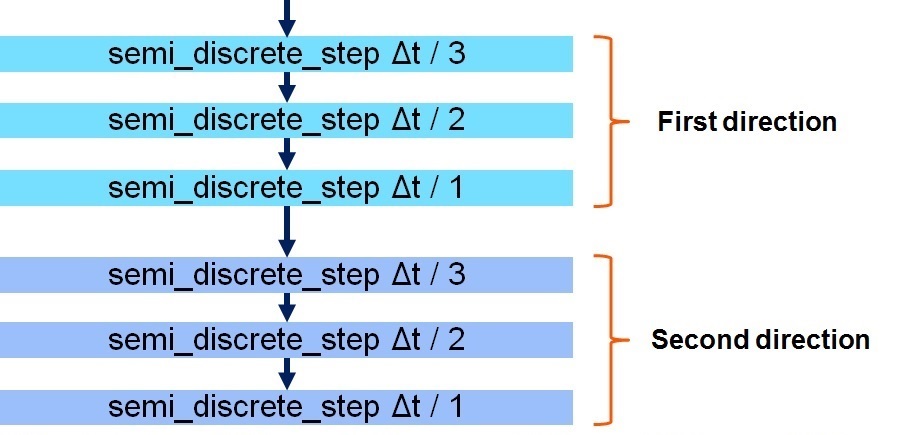
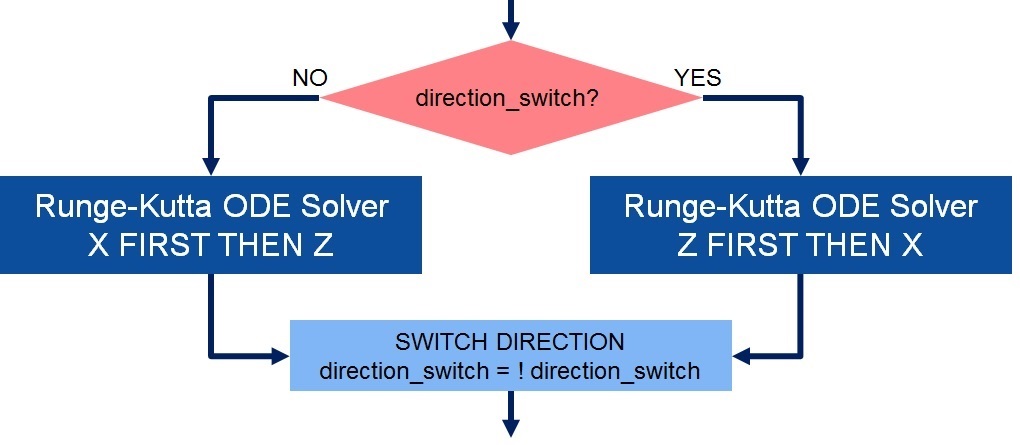
+ if (direction_switch)
|
|
|
+ {
|
|
|
+ //x-direction first
|
|
|
+ semi_discrete_step(state, state, state_tmp, dt / 3, DIR_X, flux, tend);
|
|
|
+ semi_discrete_step(state, state_tmp, state_tmp, dt / 2, DIR_X, flux, tend);
|
|
|
+ semi_discrete_step(state, state_tmp, state, dt / 1, DIR_X, flux, tend);
|
|
|
+ //z-direction second
|
|
|
+ semi_discrete_step(state, state, state_tmp, dt / 3, DIR_Z, flux, tend);
|
|
|
+ semi_discrete_step(state, state_tmp, state_tmp, dt / 2, DIR_Z, flux, tend);
|
|
|
+ semi_discrete_step(state, state_tmp, state, dt / 1, DIR_Z, flux, tend);
|
|
|
+ }
|
|
|
+ else
|
|
|
+ {
|
|
|
+ //z-direction second
|
|
|
+ semi_discrete_step(state, state, state_tmp, dt / 3, DIR_Z, flux, tend);
|
|
|
+ semi_discrete_step(state, state_tmp, state_tmp, dt / 2, DIR_Z, flux, tend);
|
|
|
+ semi_discrete_step(state, state_tmp, state, dt / 1, DIR_Z, flux, tend);
|
|
|
+ //x-direction first
|
|
|
+ semi_discrete_step(state, state, state_tmp, dt / 3, DIR_X, flux, tend);
|
|
|
+ semi_discrete_step(state, state_tmp, state_tmp, dt / 2, DIR_X, flux, tend);
|
|
|
+ semi_discrete_step(state, state_tmp, state, dt / 1, DIR_X, flux, tend);
|
|
|
+ }
|
|
|
+ if (direction_switch)
|
|
|
+ {
|
|
|
+ direction_switch = 0;
|
|
|
+ }
|
|
|
+ else
|
|
|
+ {
|
|
|
+ direction_switch = 1;
|
|
|
+ }
|
|
|
+}
|
|
|
+
|
|
|
+//Perform a single semi-discretized step in time with the form:
|
|
|
+//state_out = state_init + dt * rhs(state_forcing)
|
|
|
+//Meaning the step starts from state_init, computes the rhs using state_forcing, and stores the result in state_out
|
|
|
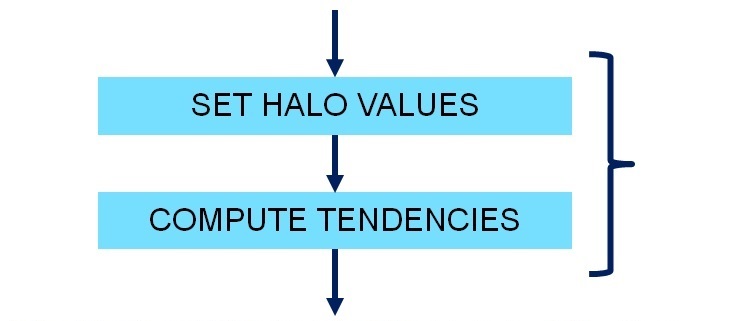
+void semi_discrete_step(double *state_init, double *state_forcing, double *state_out, double dt, int dir, double *flux, double *tend)
|
|
|
+{
|
|
|
+ int i, k, ll, inds, indt;
|
|
|
+ if (dir == DIR_X)
|
|
|
+ {
|
|
|
+ //Set the halo values in the x-direction
|
|
|
+ set_halo_values_x(state_forcing);
|
|
|
+ //Compute the time tendencies for the fluid state in the x-direction
|
|
|
+ compute_tendencies_x(state_forcing, flux, tend);
|
|
|
+ }
|
|
|
+ else if (dir == DIR_Z)
|
|
|
+ {
|
|
|
+ //Set the halo values in the z-direction
|
|
|
+ set_halo_values_z(state_forcing);
|
|
|
+ //Compute the time tendencies for the fluid state in the z-direction
|
|
|
+ compute_tendencies_z(state_forcing, flux, tend);
|
|
|
+ }
|
|
|
+
|
|
|
+/////////////////////////////////////////////////
|
|
|
+// TODO: THREAD ME
|
|
|
+/////////////////////////////////////////////////
|
|
|
+//Apply the tendencies to the fluid state
|
|
|
+#pragma acc parallel loop private(inds, indt) default(present)
|
|
|
+ for (ll = 0; ll < NUM_VARS; ll++)
|
|
|
+ {
|
|
|
+ for (k = 0; k < nz; k++)
|
|
|
+ {
|
|
|
+ for (i = 0; i < nx; i++)
|
|
|
+ {
|
|
|
+ inds = ll * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + i + hs;
|
|
|
+ indt = ll * nz * nx + k * nx + i;
|
|
|
+ state_out[inds] = state_init[inds] + dt * tend[indt];
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+}
|
|
|
+
|
|
|
+//Compute the time tendencies of the fluid state using forcing in the x-direction
|
|
|
+
|
|
|
+//First, compute the flux vector at each cell interface in the x-direction (including hyperviscosity)
|
|
|
+//Then, compute the tendencies using those fluxes
|
|
|
+void compute_tendencies_x(double *state, double *flux, double *tend)
|
|
|
+{
|
|
|
+ int i, k, ll, s, inds, indf1, indf2, indt;
|
|
|
+ double r, u, w, t, p, stencil[4], d3_vals[NUM_VARS], vals[NUM_VARS], hv_coef;
|
|
|
+ //Compute the hyperviscosity coeficient
|
|
|
+ hv_coef = -hv_beta * dx / (16 * dt);
|
|
|
+ /////////////////////////////////////////////////
|
|
|
+ // TODO: THREAD ME
|
|
|
+ /////////////////////////////////////////////////
|
|
|
+ //Compute fluxes in the x-direction for each cell
|
|
|
+#pragma acc parallel loop private(ll, s, inds, stencil, vals, d3_vals, r, u, w, t, p)
|
|
|
+ for (k = 0; k < nz; k++)
|
|
|
+ {
|
|
|
+ for (i = 0; i < nx + 1; i++)
|
|
|
+ {
|
|
|
+ //Use fourth-order interpolation from four cell averages to compute the value at the interface in question
|
|
|
+ for (ll = 0; ll < NUM_VARS; ll++)
|
|
|
+ {
|
|
|
+ for (s = 0; s < sten_size; s++)
|
|
|
+ {
|
|
|
+ inds = ll * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + i + s;
|
|
|
+ stencil[s] = state[inds];
|
|
|
+ }
|
|
|
+ //Fourth-order-accurate interpolation of the state
|
|
|
+ vals[ll] = -stencil[0] / 12 + 7 * stencil[1] / 12 + 7 * stencil[2] / 12 - stencil[3] / 12;
|
|
|
+ //First-order-accurate interpolation of the third spatial derivative of the state (for artificial viscosity)
|
|
|
+ d3_vals[ll] = -stencil[0] + 3 * stencil[1] - 3 * stencil[2] + stencil[3];
|
|
|
+ }
|
|
|
+
|
|
|
+ //Compute density, u-wind, w-wind, potential temperature, and pressure (r,u,w,t,p respectively)
|
|
|
+ r = vals[ID_DENS] + hy_dens_cell[k + hs];
|
|
|
+ u = vals[ID_UMOM] / r;
|
|
|
+ w = vals[ID_WMOM] / r;
|
|
|
+ t = (vals[ID_RHOT] + hy_dens_theta_cell[k + hs]) / r;
|
|
|
+ p = C0 * pow((r * t), gamm);
|
|
|
+
|
|
|
+ //Compute the flux vector
|
|
|
+ flux[ID_DENS * (nz + 1) * (nx + 1) + k * (nx + 1) + i] = r * u - hv_coef * d3_vals[ID_DENS];
|
|
|
+ flux[ID_UMOM * (nz + 1) * (nx + 1) + k * (nx + 1) + i] = r * u * u + p - hv_coef * d3_vals[ID_UMOM];
|
|
|
+ flux[ID_WMOM * (nz + 1) * (nx + 1) + k * (nx + 1) + i] = r * u * w - hv_coef * d3_vals[ID_WMOM];
|
|
|
+ flux[ID_RHOT * (nz + 1) * (nx + 1) + k * (nx + 1) + i] = r * u * t - hv_coef * d3_vals[ID_RHOT];
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+/////////////////////////////////////////////////
|
|
|
+// TODO: THREAD ME
|
|
|
+/////////////////////////////////////////////////
|
|
|
+//Use the fluxes to compute tendencies for each cell
|
|
|
+#pragma acc parallel loop private(indt, indf1, indf2)
|
|
|
+ for (ll = 0; ll < NUM_VARS; ll++)
|
|
|
+ {
|
|
|
+ for (k = 0; k < nz; k++)
|
|
|
+ {
|
|
|
+ for (i = 0; i < nx; i++)
|
|
|
+ {
|
|
|
+ indt = ll * nz * nx + k * nx + i;
|
|
|
+ indf1 = ll * (nz + 1) * (nx + 1) + k * (nx + 1) + i;
|
|
|
+ indf2 = ll * (nz + 1) * (nx + 1) + k * (nx + 1) + i + 1;
|
|
|
+ tend[indt] = -(flux[indf2] - flux[indf1]) / dx;
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+}
|
|
|
+
|
|
|
+//Compute the time tendencies of the fluid state using forcing in the z-direction
|
|
|
+
|
|
|
+//First, compute the flux vector at each cell interface in the z-direction (including hyperviscosity)
|
|
|
+//Then, compute the tendencies using those fluxes
|
|
|
+void compute_tendencies_z(double *state, double *flux, double *tend)
|
|
|
+{
|
|
|
+ int i, k, ll, s, inds, indf1, indf2, indt;
|
|
|
+ double r, u, w, t, p, stencil[4], d3_vals[NUM_VARS], vals[NUM_VARS], hv_coef;
|
|
|
+ //Compute the hyperviscosity coeficient
|
|
|
+ hv_coef = -hv_beta * dx / (16 * dt);
|
|
|
+/////////////////////////////////////////////////
|
|
|
+// TODO: THREAD ME
|
|
|
+/////////////////////////////////////////////////
|
|
|
+//Compute fluxes in the x-direction for each cell
|
|
|
+#pragma acc parallel loop private(ll, s, inds, stencil, vals, d3_vals, r, u, w, t, p)
|
|
|
+ for (k = 0; k < nz + 1; k++)
|
|
|
+ {
|
|
|
+ for (i = 0; i < nx; i++)
|
|
|
+ {
|
|
|
+ //Use fourth-order interpolation from four cell averages to compute the value at the interface in question
|
|
|
+ for (ll = 0; ll < NUM_VARS; ll++)
|
|
|
+ {
|
|
|
+ for (s = 0; s < sten_size; s++)
|
|
|
+ {
|
|
|
+ inds = ll * (nz + 2 * hs) * (nx + 2 * hs) + (k + s) * (nx + 2 * hs) + i + hs;
|
|
|
+ stencil[s] = state[inds];
|
|
|
+ }
|
|
|
+ //Fourth-order-accurate interpolation of the state
|
|
|
+ vals[ll] = -stencil[0] / 12 + 7 * stencil[1] / 12 + 7 * stencil[2] / 12 - stencil[3] / 12;
|
|
|
+ //First-order-accurate interpolation of the third spatial derivative of the state
|
|
|
+ d3_vals[ll] = -stencil[0] + 3 * stencil[1] - 3 * stencil[2] + stencil[3];
|
|
|
+ }
|
|
|
+
|
|
|
+ //Compute density, u-wind, w-wind, potential temperature, and pressure (r,u,w,t,p respectively)
|
|
|
+ r = vals[ID_DENS] + hy_dens_int[k];
|
|
|
+ u = vals[ID_UMOM] / r;
|
|
|
+ w = vals[ID_WMOM] / r;
|
|
|
+ t = (vals[ID_RHOT] + hy_dens_theta_int[k]) / r;
|
|
|
+ p = C0 * pow((r * t), gamm) - hy_pressure_int[k];
|
|
|
+
|
|
|
+ //Compute the flux vector with hyperviscosity
|
|
|
+ flux[ID_DENS * (nz + 1) * (nx + 1) + k * (nx + 1) + i] = r * w - hv_coef * d3_vals[ID_DENS];
|
|
|
+ flux[ID_UMOM * (nz + 1) * (nx + 1) + k * (nx + 1) + i] = r * w * u - hv_coef * d3_vals[ID_UMOM];
|
|
|
+ flux[ID_WMOM * (nz + 1) * (nx + 1) + k * (nx + 1) + i] = r * w * w + p - hv_coef * d3_vals[ID_WMOM];
|
|
|
+ flux[ID_RHOT * (nz + 1) * (nx + 1) + k * (nx + 1) + i] = r * w * t - hv_coef * d3_vals[ID_RHOT];
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+/////////////////////////////////////////////////
|
|
|
+// TODO: THREAD ME
|
|
|
+/////////////////////////////////////////////////
|
|
|
+//Use the fluxes to compute tendencies for each cell
|
|
|
+#pragma acc parallel loop private(indt, indf1, indf2)
|
|
|
+ for (ll = 0; ll < NUM_VARS; ll++)
|
|
|
+ {
|
|
|
+ for (k = 0; k < nz; k++)
|
|
|
+ {
|
|
|
+ for (i = 0; i < nx; i++)
|
|
|
+ {
|
|
|
+ indt = ll * nz * nx + k * nx + i;
|
|
|
+ indf1 = ll * (nz + 1) * (nx + 1) + (k) * (nx + 1) + i;
|
|
|
+ indf2 = ll * (nz + 1) * (nx + 1) + (k + 1) * (nx + 1) + i;
|
|
|
+ tend[indt] = -(flux[indf2] - flux[indf1]) / dz;
|
|
|
+ if (ll == ID_WMOM)
|
|
|
+ {
|
|
|
+ inds = ID_DENS * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + i + hs;
|
|
|
+ tend[indt] = tend[indt] - state[inds] * grav;
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+}
|
|
|
+
|
|
|
+void set_halo_values_x(double *state)
|
|
|
+{
|
|
|
+ int k, ll, ind_r, ind_u, ind_t, i;
|
|
|
+ double z;
|
|
|
+
|
|
|
+#pragma acc parallel loop
|
|
|
+ for (ll = 0; ll < NUM_VARS; ll++)
|
|
|
+ {
|
|
|
+ for (k = 0; k < nz; k++)
|
|
|
+ {
|
|
|
+ state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + 0] = state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + nx + hs - 2];
|
|
|
+ state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + 1] = state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + nx + hs - 1];
|
|
|
+ state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + nx + hs] = state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + hs];
|
|
|
+ state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + nx + hs + 1] = state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + hs + 1];
|
|
|
+ }
|
|
|
+ }
|
|
|
+ ////////////////////////////////////////////////////
|
|
|
+
|
|
|
+ if (myrank == 0)
|
|
|
+ {
|
|
|
+ for (k = 0; k < nz; k++)
|
|
|
+ {
|
|
|
+ for (i = 0; i < hs; i++)
|
|
|
+ {
|
|
|
+ z = (k_beg + k + 0.5) * dz;
|
|
|
+ if (abs(z - 3 * zlen / 4) <= zlen / 16)
|
|
|
+ {
|
|
|
+ ind_r = ID_DENS * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + i;
|
|
|
+ ind_u = ID_UMOM * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + i;
|
|
|
+ ind_t = ID_RHOT * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + i;
|
|
|
+ state[ind_u] = (state[ind_r] + hy_dens_cell[k + hs]) * 50.;
|
|
|
+ state[ind_t] = (state[ind_r] + hy_dens_cell[k + hs]) * 298. - hy_dens_theta_cell[k + hs];
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+}
|
|
|
+
|
|
|
+//Set this task's halo values in the z-direction.
|
|
|
+//decomposition in the vertical direction.
|
|
|
+void set_halo_values_z(double *state)
|
|
|
+{
|
|
|
+ int i, ll;
|
|
|
+ const double mnt_width = xlen / 8;
|
|
|
+ double x, xloc, mnt_deriv;
|
|
|
+/////////////////////////////////////////////////
|
|
|
+// TODO: THREAD ME
|
|
|
+/////////////////////////////////////////////////
|
|
|
+#pragma acc parallel loop private(x, xloc, mnt_deriv)
|
|
|
+ for (ll = 0; ll < NUM_VARS; ll++)
|
|
|
+ {
|
|
|
+ for (i = 0; i < nx + 2 * hs; i++)
|
|
|
+ {
|
|
|
+ if (ll == ID_WMOM)
|
|
|
+ {
|
|
|
+ state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (0) * (nx + 2 * hs) + i] = 0.;
|
|
|
+ state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (1) * (nx + 2 * hs) + i] = 0.;
|
|
|
+ state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (nz + hs) * (nx + 2 * hs) + i] = 0.;
|
|
|
+ state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (nz + hs + 1) * (nx + 2 * hs) + i] = 0.;
|
|
|
+ }
|
|
|
+ else
|
|
|
+ {
|
|
|
+ state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (0) * (nx + 2 * hs) + i] = state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (hs) * (nx + 2 * hs) + i];
|
|
|
+ state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (1) * (nx + 2 * hs) + i] = state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (hs) * (nx + 2 * hs) + i];
|
|
|
+ state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (nz + hs) * (nx + 2 * hs) + i] = state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (nz + hs - 1) * (nx + 2 * hs) + i];
|
|
|
+ state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (nz + hs + 1) * (nx + 2 * hs) + i] = state[ll * (nz + 2 * hs) * (nx + 2 * hs) + (nz + hs - 1) * (nx + 2 * hs) + i];
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+}
|
|
|
+
|
|
|
+void init()
|
|
|
+{
|
|
|
+ int i, k, ii, kk, ll, inds, i_end;
|
|
|
+ double x, z, r, u, w, t, hr, ht, nper;
|
|
|
+
|
|
|
+ //Set the cell grid size
|
|
|
+ dx = xlen / nx_glob;
|
|
|
+ dz = zlen / nz_glob;
|
|
|
+
|
|
|
+ nranks = 1;
|
|
|
+ myrank = 0;
|
|
|
+
|
|
|
+ // For simpler version, replace i_beg = 0, nx = nx_glob, left_rank = 0, right_rank = 0;
|
|
|
+
|
|
|
+ nper = ((double)nx_glob) / nranks;
|
|
|
+ i_beg = round(nper * (myrank));
|
|
|
+ i_end = round(nper * ((myrank) + 1)) - 1;
|
|
|
+ nx = i_end - i_beg + 1;
|
|
|
+ left_rank = myrank - 1;
|
|
|
+ if (left_rank == -1)
|
|
|
+ left_rank = nranks - 1;
|
|
|
+ right_rank = myrank + 1;
|
|
|
+ if (right_rank == nranks)
|
|
|
+ right_rank = 0;
|
|
|
+
|
|
|
+ ////////////////////////////////////////////////////////////////////////////////
|
|
|
+ ////////////////////////////////////////////////////////////////////////////////
|
|
|
+ // YOU DON'T NEED TO ALTER ANYTHING BELOW THIS POINT IN THE CODE
|
|
|
+ ////////////////////////////////////////////////////////////////////////////////
|
|
|
+ ////////////////////////////////////////////////////////////////////////////////
|
|
|
+
|
|
|
+ k_beg = 0;
|
|
|
+ nz = nz_glob;
|
|
|
+
|
|
|
+ //Allocate the model data
|
|
|
+ state = (double *)malloc((nx + 2 * hs) * (nz + 2 * hs) * NUM_VARS * sizeof(double));
|
|
|
+ state_tmp = (double *)malloc((nx + 2 * hs) * (nz + 2 * hs) * NUM_VARS * sizeof(double));
|
|
|
+ flux = (double *)malloc((nx + 1) * (nz + 1) * NUM_VARS * sizeof(double));
|
|
|
+ tend = (double *)malloc(nx * nz * NUM_VARS * sizeof(double));
|
|
|
+ hy_dens_cell = (double *)malloc((nz + 2 * hs) * sizeof(double));
|
|
|
+ hy_dens_theta_cell = (double *)malloc((nz + 2 * hs) * sizeof(double));
|
|
|
+ hy_dens_int = (double *)malloc((nz + 1) * sizeof(double));
|
|
|
+ hy_dens_theta_int = (double *)malloc((nz + 1) * sizeof(double));
|
|
|
+ hy_pressure_int = (double *)malloc((nz + 1) * sizeof(double));
|
|
|
+
|
|
|
+ //Define the maximum stable time step based on an assumed maximum wind speed
|
|
|
+ dt = dmin(dx, dz) / max_speed * cfl;
|
|
|
+ //Set initial elapsed model time and output_counter to zero
|
|
|
+ etime = 0.;
|
|
|
+ output_counter = 0.;
|
|
|
+
|
|
|
+ // Display grid information
|
|
|
+
|
|
|
+ printf("nx_glob, nz_glob: %d %d\n", nx_glob, nz_glob);
|
|
|
+ printf("dx,dz: %lf %lf\n", dx, dz);
|
|
|
+ printf("dt: %lf\n", dt);
|
|
|
+
|
|
|
+ //////////////////////////////////////////////////////////////////////////
|
|
|
+ // Initialize the cell-averaged fluid state via Gauss-Legendre quadrature
|
|
|
+ //////////////////////////////////////////////////////////////////////////
|
|
|
+ for (k = 0; k < nz + 2 * hs; k++)
|
|
|
+ {
|
|
|
+ for (i = 0; i < nx + 2 * hs; i++)
|
|
|
+ {
|
|
|
+ //Initialize the state to zero
|
|
|
+ for (ll = 0; ll < NUM_VARS; ll++)
|
|
|
+ {
|
|
|
+ inds = ll * (nz + 2 * hs) * (nx + 2 * hs) + k * (nx + 2 * hs) + i;
|
|
|
+ state[inds] = 0.;
|
|
|
+ }
|
|
|
+ //Use Gauss-Legendre quadrature to initialize a hydrostatic balance + temperature perturbation
|
|
|
+ for (kk = 0; kk < nqpoints; kk++)
|
|
|
+ {
|
|
|
+ for (ii = 0; ii < nqpoints; ii++)
|
|
|
+ {
|
|
|
+ //Compute the x,z location within the global domain based on cell and quadrature index
|
|
|
+ x = (i_beg + i - hs + 0.5) * dx + (qpoints[ii] - 0.5) * dx;
|
|
|
+ z = (k_beg + k - hs + 0.5) * dz + (qpoints[kk] - 0.5) * dz;
|
|
|
+
|
|
|
+ //Set the fluid state based on the user's specification (default is injection in this example)
|
|
|
+ injection(x, z, r, u, w, t, hr, ht);
|
|
|
+
|
|
|
+ //Store into the fluid state array
|
|
|
+ inds = ID_DENS * (nz + 2 * hs) * (nx + 2 * hs) + k * (nx + 2 * hs) + i;
|
|
|
+ state[inds] = state[inds] + r * qweights[ii] * qweights[kk];
|
|
|
+ inds = ID_UMOM * (nz + 2 * hs) * (nx + 2 * hs) + k * (nx + 2 * hs) + i;
|
|
|
+ state[inds] = state[inds] + (r + hr) * u * qweights[ii] * qweights[kk];
|
|
|
+ inds = ID_WMOM * (nz + 2 * hs) * (nx + 2 * hs) + k * (nx + 2 * hs) + i;
|
|
|
+ state[inds] = state[inds] + (r + hr) * w * qweights[ii] * qweights[kk];
|
|
|
+ inds = ID_RHOT * (nz + 2 * hs) * (nx + 2 * hs) + k * (nx + 2 * hs) + i;
|
|
|
+ state[inds] = state[inds] + ((r + hr) * (t + ht) - hr * ht) * qweights[ii] * qweights[kk];
|
|
|
+ }
|
|
|
+ }
|
|
|
+ for (ll = 0; ll < NUM_VARS; ll++)
|
|
|
+ {
|
|
|
+ inds = ll * (nz + 2 * hs) * (nx + 2 * hs) + k * (nx + 2 * hs) + i;
|
|
|
+ state_tmp[inds] = state[inds];
|
|
|
+ }
|
|
|
+ }
|
|
|
+ }
|
|
|
+ //Compute the hydrostatic background state over vertical cell averages
|
|
|
+ for (k = 0; k < nz + 2 * hs; k++)
|
|
|
+ {
|
|
|
+ hy_dens_cell[k] = 0.;
|
|
|
+ hy_dens_theta_cell[k] = 0.;
|
|
|
+ for (kk = 0; kk < nqpoints; kk++)
|
|
|
+ {
|
|
|
+ z = (k_beg + k - hs + 0.5) * dz;
|
|
|
+
|
|
|
+ //Set the fluid state based on the user's specification (default is injection in this example)
|
|
|
+ injection(0., z, r, u, w, t, hr, ht);
|
|
|
+
|
|
|
+ hy_dens_cell[k] = hy_dens_cell[k] + hr * qweights[kk];
|
|
|
+ hy_dens_theta_cell[k] = hy_dens_theta_cell[k] + hr * ht * qweights[kk];
|
|
|
+ }
|
|
|
+ }
|
|
|
+ //Compute the hydrostatic background state at vertical cell interfaces
|
|
|
+ for (k = 0; k < nz + 1; k++)
|
|
|
+ {
|
|
|
+ z = (k_beg + k) * dz;
|
|
|
+
|
|
|
+ //Set the fluid state based on the user's specification (default is injection in this example)
|
|
|
+ injection(0., z, r, u, w, t, hr, ht);
|
|
|
+
|
|
|
+ hy_dens_int[k] = hr;
|
|
|
+ hy_dens_theta_int[k] = hr * ht;
|
|
|
+ hy_pressure_int[k] = C0 * pow((hr * ht), gamm);
|
|
|
+ }
|
|
|
+}
|
|
|
+
|
|
|
+//This test case is initially balanced but injects fast, cold air from the left boundary near the model top
|
|
|
+//x and z are input coordinates at which to sample
|
|
|
+//r,u,w,t are output density, u-wind, w-wind, and potential temperature at that location
|
|
|
+//hr and ht are output background hydrostatic density and potential temperature at that location
|
|
|
+void injection(double x, double z, double &r, double &u, double &w, double &t, double &hr, double &ht)
|
|
|
+{
|
|
|
+ hydro_const_theta(z, hr, ht);
|
|
|
+ r = 0.;
|
|
|
+ t = 0.;
|
|
|
+ u = 0.;
|
|
|
+ w = 0.;
|
|
|
+}
|
|
|
+
|
|
|
+//Establish hydrstatic balance using constant potential temperature (thermally neutral atmosphere)
|
|
|
+//z is the input coordinate
|
|
|
+//r and t are the output background hydrostatic density and potential temperature
|
|
|
+void hydro_const_theta(double z, double &r, double &t)
|
|
|
+{
|
|
|
+ const double theta0 = 300.; //Background potential temperature
|
|
|
+ const double exner0 = 1.; //Surface-level Exner pressure
|
|
|
+ double p, exner, rt;
|
|
|
+ //Establish hydrostatic balance first using Exner pressure
|
|
|
+ t = theta0; //Potential Temperature at z
|
|
|
+ exner = exner0 - grav * z / (cp * theta0); //Exner pressure at z
|
|
|
+ p = p0 * pow(exner, (cp / rd)); //Pressure at z
|
|
|
+ rt = pow((p / C0), (1. / gamm)); //rho*theta at z
|
|
|
+ r = rt / t; //Density at z
|
|
|
+}
|
|
|
+
|
|
|
+//Output the fluid state (state) to a NetCDF file at a given elapsed model time (etime)
|
|
|
+//The file I/O uses netcdf, the only external library required for this mini-app.
|
|
|
+//If it's too cumbersome, you can comment the I/O out, but you'll miss out on some potentially cool graphics
|
|
|
+void output(double *state, double etime)
|
|
|
+{
|
|
|
+ int ncid, t_dimid, x_dimid, z_dimid, dens_varid, uwnd_varid, wwnd_varid, theta_varid, t_varid, dimids[3];
|
|
|
+ int i, k, ind_r, ind_u, ind_w, ind_t;
|
|
|
+
|
|
|
+ size_t st1[1], ct1[1], st3[3], ct3[3];
|
|
|
+
|
|
|
+ //Temporary arrays to hold density, u-wind, w-wind, and potential temperature (theta)
|
|
|
+ double *dens, *uwnd, *wwnd, *theta;
|
|
|
+ double *etimearr;
|
|
|
+ //Inform the user
|
|
|
+
|
|
|
+ printf("*** OUTPUT ***\n");
|
|
|
+
|
|
|
+ //Allocate some (big) temp arrays
|
|
|
+ dens = (double *)malloc(nx * nz * sizeof(double));
|
|
|
+ uwnd = (double *)malloc(nx * nz * sizeof(double));
|
|
|
+ wwnd = (double *)malloc(nx * nz * sizeof(double));
|
|
|
+ theta = (double *)malloc(nx * nz * sizeof(double));
|
|
|
+ etimearr = (double *)malloc(1 * sizeof(double));
|
|
|
+
|
|
|
+ //If the elapsed time is zero, create the file. Otherwise, open the file
|
|
|
+ if (etime == 0)
|
|
|
+ {
|
|
|
+ //Create the file
|
|
|
+ ncwrap(nc_create("new.nc", NC_CLOBBER, &ncid), __LINE__);
|
|
|
+
|
|
|
+ //Create the dimensions
|
|
|
+ ncwrap(nc_def_dim(ncid, "t", NC_UNLIMITED, &t_dimid), __LINE__);
|
|
|
+ ncwrap(nc_def_dim(ncid, "x", nx_glob, &x_dimid), __LINE__);
|
|
|
+ ncwrap(nc_def_dim(ncid, "z", nz_glob, &z_dimid), __LINE__);
|
|
|
+
|
|
|
+ //Create the variables
|
|
|
+ dimids[0] = t_dimid;
|
|
|
+ ncwrap(nc_def_var(ncid, "t", NC_DOUBLE, 1, dimids, &t_varid), __LINE__);
|
|
|
+
|
|
|
+ dimids[0] = t_dimid;
|
|
|
+ dimids[1] = z_dimid;
|
|
|
+ dimids[2] = x_dimid;
|
|
|
+
|
|
|
+ ncwrap(nc_def_var(ncid, "dens", NC_DOUBLE, 3, dimids, &dens_varid), __LINE__);
|
|
|
+ ncwrap(nc_def_var(ncid, "uwnd", NC_DOUBLE, 3, dimids, &uwnd_varid), __LINE__);
|
|
|
+ ncwrap(nc_def_var(ncid, "wwnd", NC_DOUBLE, 3, dimids, &wwnd_varid), __LINE__);
|
|
|
+ ncwrap(nc_def_var(ncid, "theta", NC_DOUBLE, 3, dimids, &theta_varid), __LINE__);
|
|
|
+
|
|
|
+ //End "define" mode
|
|
|
+ ncwrap(nc_enddef(ncid), __LINE__);
|
|
|
+ }
|
|
|
+ else
|
|
|
+ {
|
|
|
+ //Open the file
|
|
|
+ ncwrap(nc_open("new.nc", NC_WRITE, &ncid), __LINE__);
|
|
|
+
|
|
|
+ //Get the variable IDs
|
|
|
+ ncwrap(nc_inq_varid(ncid, "dens", &dens_varid), __LINE__);
|
|
|
+ ncwrap(nc_inq_varid(ncid, "uwnd", &uwnd_varid), __LINE__);
|
|
|
+ ncwrap(nc_inq_varid(ncid, "wwnd", &wwnd_varid), __LINE__);
|
|
|
+ ncwrap(nc_inq_varid(ncid, "theta", &theta_varid), __LINE__);
|
|
|
+ ncwrap(nc_inq_varid(ncid, "t", &t_varid), __LINE__);
|
|
|
+ }
|
|
|
+
|
|
|
+ //Store perturbed values in the temp arrays for output
|
|
|
+ for (k = 0; k < nz; k++)
|
|
|
+ {
|
|
|
+ for (i = 0; i < nx; i++)
|
|
|
+ {
|
|
|
+ ind_r = ID_DENS * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + i + hs;
|
|
|
+ ind_u = ID_UMOM * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + i + hs;
|
|
|
+ ind_w = ID_WMOM * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + i + hs;
|
|
|
+ ind_t = ID_RHOT * (nz + 2 * hs) * (nx + 2 * hs) + (k + hs) * (nx + 2 * hs) + i + hs;
|
|
|
+ dens[k * nx + i] = state[ind_r];
|
|
|
+ uwnd[k * nx + i] = state[ind_u] / (hy_dens_cell[k + hs] + state[ind_r]);
|
|
|
+ wwnd[k * nx + i] = state[ind_w] / (hy_dens_cell[k + hs] + state[ind_r]);
|
|
|
+ theta[k * nx + i] = (state[ind_t] + hy_dens_theta_cell[k + hs]) / (hy_dens_cell[k + hs] + state[ind_r]) - hy_dens_theta_cell[k + hs] / hy_dens_cell[k + hs];
|
|
|
+ }
|
|
|
+ }
|
|
|
+
|
|
|
+ //Write the grid data to file with all the processes writing collectively
|
|
|
+ st3[0] = num_out;
|
|
|
+ st3[1] = k_beg;
|
|
|
+ st3[2] = i_beg;
|
|
|
+ ct3[0] = 1;
|
|
|
+ ct3[1] = nz;
|
|
|
+ ct3[2] = nx;
|
|
|
+
|
|
|
+ ncwrap(nc_put_vara_double(ncid, dens_varid, st3, ct3, dens), __LINE__);
|
|
|
+ ncwrap(nc_put_vara_double(ncid, uwnd_varid, st3, ct3, uwnd), __LINE__);
|
|
|
+ ncwrap(nc_put_vara_double(ncid, wwnd_varid, st3, ct3, wwnd), __LINE__);
|
|
|
+ ncwrap(nc_put_vara_double(ncid, theta_varid, st3, ct3, theta), __LINE__);
|
|
|
+
|
|
|
+ //Only the master process needs to write the elapsed time
|
|
|
+ //write elapsed time to file
|
|
|
+
|
|
|
+ st1[0] = num_out;
|
|
|
+ ct1[0] = 1;
|
|
|
+ etimearr[0] = etime;
|
|
|
+ ncwrap(nc_put_vara_double(ncid, t_varid, st1, ct1, etimearr), __LINE__);
|
|
|
+
|
|
|
+ //Close the file
|
|
|
+ ncwrap(nc_close(ncid), __LINE__);
|
|
|
+
|
|
|
+ //Increment the number of outputs
|
|
|
+ num_out = num_out + 1;
|
|
|
+
|
|
|
+ //Deallocate the temp arrays
|
|
|
+ free(dens);
|
|
|
+ free(uwnd);
|
|
|
+ free(wwnd);
|
|
|
+ free(theta);
|
|
|
+ free(etimearr);
|
|
|
+}
|
|
|
+
|
|
|
+//Error reporting routine for the NetCDF I/O
|
|
|
+void ncwrap(int ierr, int line)
|
|
|
+{
|
|
|
+ if (ierr != NC_NOERR)
|
|
|
+ {
|
|
|
+ printf("NetCDF Error at line: %d\n", line);
|
|
|
+ printf("%s\n", nc_strerror(ierr));
|
|
|
+ exit(-1);
|
|
|
+ }
|
|
|
+}
|
|
|
+
|
|
|
+void finalize()
|
|
|
+{
|
|
|
+ free(state);
|
|
|
+ free(state_tmp);
|
|
|
+ free(flux);
|
|
|
+ free(tend);
|
|
|
+ free(hy_dens_cell);
|
|
|
+ free(hy_dens_theta_cell);
|
|
|
+ free(hy_dens_int);
|
|
|
+ free(hy_dens_theta_int);
|
|
|
+ free(hy_pressure_int);
|
|
|
+}
|